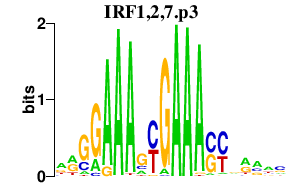
|
chr6_+_121195885
|
260.099
|
NM_011909
|
Usp18
|
ubiquitin specific peptidase 18
|
|
chr5_-_121357467
|
210.385
|
NM_145211
|
Oas1a
|
2'-5' oligoadenylate synthetase 1A
|
|
chr4_-_155574875
|
188.343
|
NM_015783
|
Isg15
|
ISG15 ubiquitin-like modifier
|
|
chr1_-_175465858
|
180.573
|
|
BC094916
|
cDNA sequence BC094916
|
|
chr3_+_142283605
|
168.413
|
NM_010260
|
Gbp2
|
guanylate binding protein 2
|
|
chr5_-_121337533
|
166.687
|
NM_011852
|
Oas1g
|
2'-5' oligoadenylate synthetase 1G
|
|
chr1_-_175810248
|
161.412
|
NM_001170853
|
Mndal
|
myeloid nuclear differentiation antigen like
|
|
chr11_+_48900507
|
156.137
|
NM_008330
|
Ifi47
Olfr56
|
interferon gamma inducible protein 47
olfactory receptor 56
|
|
chr6_+_57531124
|
155.955
|
|
Herc6
|
hect domain and RLD 6
|
|
chr19_+_34658016
|
155.756
|
NM_010501
|
Ifit3
|
interferon-induced protein with tetratricopeptide repeats 3
|
|
chr16_+_23609986
|
152.536
|
NM_023386
|
Rtp4
|
receptor transporter protein 4
|
|
chr7_-_148452299
|
146.294
|
NM_016850
|
Irf7
|
interferon regulatory factor 7
|
|
chr17_-_34324234
|
141.784
|
NM_013585
|
Psmb9
|
proteasome (prosome, macropain) subunit, beta type 9 (large multifunctional peptidase 2)
|
|
chr19_-_11124897
|
136.766
|
NM_001177351
|
AW112010
|
expressed sequence AW112010
|
|
chr19_+_34682418
|
134.103
|
NM_001005858
|
I830012O16Rik
|
RIKEN cDNA I830012O16 gene
|
|
chr8_-_108462291
|
133.754
|
NM_013640
|
Psmb10
|
proteasome (prosome, macropain) subunit, beta type 10
|
|
chr11_-_48877704
|
133.166
|
NM_001145164
|
Tgtp2
|
T-cell specific GTPase 2
|
|
chr6_+_57530968
|
132.062
|
NM_025992
|
Herc6
|
hect domain and RLD 6
|
|
chr11_+_58013057
|
129.284
|
NM_018738
|
Igtp
|
interferon gamma induced GTPase
|
|
chr11_-_48877544
|
128.528
|
|
Tgtp2
Tgtp1
|
T-cell specific GTPase 2
T-cell specific GTPase 1
|
|
chr18_+_60542336
|
127.268
|
|
Iigp1
|
interferon inducible GTPase 1
|
|
chr2_-_173044305
|
126.806
|
NM_001139519
NM_021394
|
Zbp1
|
Z-DNA binding protein 1
|
|
chr17_+_34324700
|
125.765
|
|
Tap1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP)
|
|
chr2_+_121973414
|
123.730
|
NM_009735
|
B2m
|
beta-2 microglobulin
|
|
chrX_-_9046362
|
123.077
|
NM_007807
|
Cybb
|
cytochrome b-245, beta polypeptide
|
|
chr16_+_35939047
|
120.105
|
|
Parp9
|
poly (ADP-ribose) polymerase family, member 9
|
|
chr9_-_88626613
|
119.348
|
NM_007536
|
Bcl2a1d
|
B-cell leukemia/lymphoma 2 related protein A1d
|
|
chr10_-_33847745
|
119.310
|
NM_175449
|
Fam26f
|
family with sequence similarity 26, member F
|
|
chr17_+_34324636
|
118.782
|
|
Tap1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP)
|
|
chr11_-_78798332
|
112.710
|
NM_001159301
NM_010708
|
Lgals9
|
lectin, galactose binding, soluble 9
|
|
chr19_+_34715368
|
112.651
|
NM_008331
|
Ifit1
|
interferon-induced protein with tetratricopeptide repeats 1
|
|
chr8_-_74061134
|
112.620
|
NM_198095
|
Bst2
|
bone marrow stromal cell antigen 2
|
|
chr8_-_108461909
|
112.346
|
|
Psmb10
|
proteasome (prosome, macropain) subunit, beta type 10
|
|
chr11_-_70273251
|
112.206
|
|
Cxcl16
|
chemokine (C-X-C motif) ligand 16
|
|
chr11_-_70273190
|
108.256
|
|
Cxcl16
|
chemokine (C-X-C motif) ligand 16
|
|
chr11_-_48805585
|
108.184
|
NM_001145164
NM_011579
|
Tgtp2
Tgtp1
Gm12185
|
T-cell specific GTPase 2
T-cell specific GTPase 1
predicted gene 12185
|
|
chr11_-_70272909
|
107.749
|
|
Cxcl16
|
chemokine (C-X-C motif) ligand 16
|
|
chr5_-_92777875
|
107.556
|
NM_021274
|
Cxcl10
|
chemokine (C-X-C motif) ligand 10
|
|
chr19_+_29441927
|
107.475
|
NM_021893
|
Cd274
|
CD274 antigen
|
|
chr11_-_70273434
|
107.255
|
|
Cxcl16
|
chemokine (C-X-C motif) ligand 16
|
|
chr8_-_113862056
|
106.020
|
|
Mlkl
|
mixed lineage kinase domain-like
|
|
chr8_+_64406883
|
105.593
|
NM_001081215
|
Ddx60
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 60
|
|
chr5_-_105668551
|
102.011
|
NM_001039646
|
Gbp10
|
guanylate-binding protein 10
|
|
chr2_-_51828440
|
101.327
|
|
Nmi
|
N-myc (and STAT) interactor
|
|
chr19_+_34644768
|
100.347
|
|
Ifit2
|
interferon-induced protein with tetratricopeptide repeats 2
|
|
chr9_-_88626434
|
99.231
|
|
Bcl2a1d
|
B-cell leukemia/lymphoma 2 related protein A1d
|
|
chr14_+_103446193
|
98.897
|
|
Irg1
|
immunoresponsive gene 1
|
|
chr3_+_142193286
|
98.617
|
NM_001083312
NM_145545
|
Gbp6
|
guanylate binding protein 6
|
|
chr9_+_5298516
|
98.307
|
NM_009807
|
Casp1
|
caspase 1
|
|
chr1_-_175842977
|
97.888
|
NM_001033450
|
Ifi204
Mnda
|
interferon activated gene 204
myeloid cell nuclear differentiation antigen
|
|
chr6_-_38304508
|
97.756
|
|
Zc3hav1
|
zinc finger CCCH type, antiviral 1
|
|
chr11_-_70273333
|
96.961
|
|
Cxcl16
|
chemokine (C-X-C motif) ligand 16
|
|
chr1_+_87546558
|
96.433
|
NM_013673
|
Sp100
|
nuclear antigen Sp100
|
|
chr16_-_35939034
|
96.430
|
NM_001013371
|
Dtx3l
|
deltex 3-like (Drosophila)
|
|
chr5_-_134705423
|
96.293
|
NM_010876
|
Ncf1
|
neutrophil cytosolic factor 1
|
|
chr17_+_34335139
|
96.177
|
NM_010724
|
Psmb8
|
proteasome (prosome, macropain) subunit, beta type 8 (large multifunctional peptidase 7)
|
|
chr19_+_29446941
|
95.975
|
|
Cd274
|
CD274 antigen
|
|
chr5_-_121227617
|
95.386
|
NM_145226
|
Oas3
|
2'-5' oligoadenylate synthetase 3
|
|
chr11_+_101443690
|
94.659
|
|
Tmem106a
|
transmembrane protein 106A
|
|
chr19_+_12873063
|
94.030
|
NM_134152
|
Lpxn
|
leupaxin
|
|
chr5_+_115347505
|
93.641
|
|
Oasl2
|
2'-5' oligoadenylate synthetase-like 2
|
|
chr3_+_142223027
|
93.343
|
|
Gbp3
|
guanylate binding protein 3
|
|
chr2_-_62484264
|
92.735
|
NM_001164477
NM_027835
|
Ifih1
|
interferon induced with helicase C domain 1
|
|
chr7_-_109713935
|
91.976
|
NM_001082552
NM_009277
|
Trim21
|
tripartite motif-containing 21
|
|
chr11_-_82834223
|
90.921
|
NM_001167743
NM_181545
|
Slfn8
|
schlafen 8
|
|
chr7_-_111656285
|
89.860
|
NM_001167828
NM_199146
|
Trim30a
Trim30d
|
tripartite motif-containing 30A
tripartite motif-containing 30D
|
|
chr15_+_75692745
|
89.502
|
NM_026960
|
Gsdmd
|
gasdermin D
|
|
chr14_+_78304045
|
89.069
|
NM_029495
NM_178825
|
Epsti1
|
epithelial stromal interaction 1 (breast)
|
|
chr11_-_70273453
|
88.834
|
NM_023158
|
Cxcl16
|
chemokine (C-X-C motif) ligand 16
|
|
chr1_-_175872522
|
88.440
|
NM_008328
NM_001045481
|
Ifi203
|
interferon activated gene 203
|
|
chr13_-_100972022
|
86.322
|
NM_001126182
NM_010872
|
Naip2
|
NLR family, apoptosis inhibitory protein 2
|
|
chr14_+_103446209
|
84.314
|
NM_008392
|
Irg1
|
immunoresponsive gene 1
|
|
chr6_-_3349415
|
84.259
|
|
Samd9l
|
sterile alpha motif domain containing 9-like
|
|
chr8_-_84927109
|
84.169
|
|
Il15
|
interleukin 15
|
|
chr17_+_34324496
|
82.580
|
NM_001161730
NM_013683
|
Tap1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP)
|
|
chr11_-_48684659
|
82.433
|
NM_008326
|
Irgm1
|
immunity-related GTPase family M member 1
|
|
chr1_-_175696989
|
82.348
|
NM_008329
|
Ifi204
|
interferon activated gene 204
|
|
chr19_-_34673889
|
82.342
|
NM_001110517
|
Gm14446
|
predicted gene 14446
|
|
chr17_+_57498108
|
82.284
|
NM_010130
|
Emr1
|
EGF-like module containing, mucin-like, hormone receptor-like sequence 1
|
|
chr5_-_105722663
|
82.138
|
NM_194336
|
Mpa2l
Gbp4
|
macrophage activation 2 like
guanylate binding protein 4
|
|
chr3_+_142223007
|
82.008
|
NM_018734
|
Gbp3
|
guanylate binding protein 3
|
|
chr1_-_175671849
|
81.880
|
|
AI607873
|
expressed sequence AI607873
|
|
chr13_-_101017247
|
80.681
|
NM_010870
|
Naip5
|
NLR family, apoptosis inhibitory protein 5
|
|
chr6_+_29479250
|
80.446
|
|
Irf5
|
interferon regulatory factor 5
|
|
chr5_+_86500770
|
80.174
|
NM_019992
|
Stap1
|
signal transducing adaptor family member 1
|
|
chr14_-_59916342
|
80.068
|
NM_172603
|
Phf11
|
PHD finger protein 11
|
|
chr13_+_37437194
|
79.870
|
NM_010745
|
Ly86
|
lymphocyte antigen 86
|
|
chr1_+_175603760
|
79.369
|
NM_001162938
|
Pydc3
|
pyrin domain containing 3
|
|
chr19_+_12873115
|
78.669
|
|
Lpxn
|
leupaxin
|
|
chr1_+_52176281
|
78.318
|
NM_009283
|
Stat1
|
signal transducer and activator of transcription 1
|
|
chr6_-_3295980
|
77.492
|
|
9530028C05
|
hypothetical protein 9530028C05
|
|
chr17_-_36157472
|
76.949
|
NM_008207
|
H2-T24
|
histocompatibility 2, T region locus 24
|
|
chr7_+_111392968
|
76.456
|
NM_030684
|
Trim34a
|
tripartite motif-containing 34A
|
|
chr14_-_59984246
|
76.152
|
NM_199015
|
D14Ertd668e
|
DNA segment, Chr 14, ERATO Doi 668, expressed
|
|
chr19_+_56471731
|
74.804
|
|
Casp7
|
caspase 7
|
|
chr17_+_34341467
|
74.516
|
|
Tap2
|
transporter 2, ATP-binding cassette, sub-family B (MDR/TAP)
|
|
chr18_+_60963495
|
73.792
|
NM_001042605
NM_010545
|
Cd74
|
CD74 antigen (invariant polypeptide of major histocompatibility complex, class II antigen-associated)
|
|
chr11_+_101443529
|
73.584
|
NM_144830
|
Tmem106a
|
transmembrane protein 106A
|
|
chr11_+_101309725
|
73.421
|
NM_027320
|
Ifi35
|
interferon-induced protein 35
|
|
chr19_-_34676457
|
73.302
|
NM_001101605
|
Gm14446
|
predicted gene 14446
|
|
chr19_+_56471580
|
72.825
|
NM_007611
|
Casp7
|
caspase 7
|
|
chr3_-_151412812
|
72.602
|
NM_133871
|
Ifi44
|
interferon-induced protein 44
|
|
chr2_-_51790671
|
72.353
|
|
Rbm43
|
RNA binding motif protein 43
|
|
chr17_+_34341417
|
72.039
|
NM_011530
|
Tap2
|
transporter 2, ATP-binding cassette, sub-family B (MDR/TAP)
|
|
chr17_+_34056367
|
71.286
|
NM_001025313
NM_009318
|
Tapbp
|
TAP binding protein
|
|
chr6_-_39067918
|
71.131
|
|
Parp12
|
poly (ADP-ribose) polymerase family, member 12
|
|
chr15_+_78156227
|
70.788
|
NM_007780
|
Csf2rb
|
colony stimulating factor 2 receptor, beta, low-affinity (granulocyte-macrophage)
|
|
chr16_-_97684510
|
70.656
|
NM_010846
|
Mx1
|
myxovirus (influenza virus) resistance 1
|
|
chr2_-_51790406
|
70.502
|
NM_001141981
NM_030243
|
Rbm43
|
RNA binding motif protein 43
|
|
chr8_-_84926440
|
70.362
|
NM_008357
|
Il15
|
interleukin 15
|
|
chr9_-_110959315
|
69.438
|
|
Ccrl2
|
chemokine (C-C motif) receptor-like 2
|
|
chr10_-_87819740
|
68.317
|
|
Dram1
|
DNA-damage regulated autophagy modulator 1
|
|
chr6_-_38304558
|
67.379
|
NM_028421
NM_028864
|
Zc3hav1
|
zinc finger CCCH type, antiviral 1
|
|
chr6_-_39068241
|
67.219
|
|
Parp12
|
poly (ADP-ribose) polymerase family, member 12
|
|
chr15_-_76073582
|
66.320
|
|
Parp10
|
poly (ADP-ribose) polymerase family, member 10
|
|
chr1_+_173319048
|
66.115
|
NM_001005508
|
Arhgap30
|
Rho GTPase activating protein 30
|
|
chr15_-_76073789
|
65.547
|
|
Parp10
|
poly (ADP-ribose) polymerase family, member 10
|
|
chr17_+_34056798
|
65.523
|
|
Tapbp
|
TAP binding protein
|
|
chr4_-_40186722
|
65.248
|
NM_172689
|
Ddx58
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 58
|
|
chr5_+_115347750
|
65.184
|
|
Oasl2
|
2'-5' oligoadenylate synthetase-like 2
|
|
chr15_-_76073869
|
64.852
|
NM_001163575
NM_001163576
|
Parp10
|
poly (ADP-ribose) polymerase family, member 10
|
|
chr9_-_37063214
|
64.594
|
NM_001145960
NM_020258
|
Slc37a2
|
solute carrier family 37 (glycerol-3-phosphate transporter), member 2
|
|
chr8_-_113861802
|
64.390
|
NM_029005
|
Mlkl
|
mixed lineage kinase domain-like
|
|
chr17_+_34056555
|
64.077
|
|
Tapbp
|
TAP binding protein
|
|
chr9_-_110959702
|
63.662
|
NM_017466
|
Ccrl2
|
chemokine (C-C motif) receptor-like 2
|
|
chrX_+_106392094
|
63.285
|
NM_001037917
|
Gm6377
|
predicted gene 6377
|
|
chr17_+_34056585
|
62.846
|
|
Tapbp
|
TAP binding protein
|
|
chr15_-_76073854
|
62.812
|
|
Parp10
|
poly (ADP-ribose) polymerase family, member 10
|
|
chr1_+_58859345
|
62.778
|
NM_001080126
|
Casp8
|
caspase 8
|
|
chr19_-_3905229
|
62.385
|
NM_016921
|
Tcirg1
|
T-cell, immune regulator 1, ATPase, H+ transporting, lysosomal V0 protein A3
|
|
chr8_-_125075222
|
62.248
|
NM_001037298
|
Fam38a
|
family with sequence similarity 38, member A
|
|
chr1_-_175961813
|
60.981
|
NM_172648
|
Ifi205
Ifi202b
|
interferon activated gene 205
interferon activated gene 202B
|
|
chr15_+_83393975
|
60.949
|
NM_009775
|
Tspo
|
translocator protein
|
|
chr5_-_121199781
|
60.834
|
NM_145227
|
Oas2
|
2'-5' oligoadenylate synthetase 2
|
|
chr9_+_107877887
|
60.479
|
NM_023738
|
Uba7
|
ubiquitin-like modifier activating enzyme 7
|
|
chr10_-_94407211
|
59.951
|
NM_018797
|
Plxnc1
|
plexin C1
|
|
chr12_-_27129497
|
59.173
|
|
Rsad2
|
radical S-adenosyl methionine domain containing 2
|
|
chr5_-_105539282
|
57.797
|
NM_172777
|
Gbp9
|
guanylate-binding protein 9
|
|
chr6_+_122902532
|
57.111
|
NM_153197
|
Clec4a3
|
C-type lectin domain family 4, member a3
|
|
chr13_-_113891176
|
56.975
|
NM_010370
|
Gzma
|
granzyme A
|
|
chr5_-_105482574
|
56.922
|
NM_029509
|
Gbp8
|
guanylate-binding protein 8
|
|
chr8_-_46418352
|
56.442
|
NM_133969
|
Cyp4v3
|
cytochrome P450, family 4, subfamily v, polypeptide 3
|
|
chr17_-_34999068
|
56.271
|
|
Cfb
|
complement factor B
|
|
chr7_+_86058817
|
55.889
|
|
Isg20
|
interferon-stimulated protein
|
|
chr17_-_36258349
|
55.816
|
NM_010395
|
H2-T10
H2-T9
|
histocompatibility 2, T region locus 10
histocompatibility 2, T region locus 9
|
|
chr5_+_115346942
|
55.355
|
NM_011854
|
Oasl2
|
2'-5' oligoadenylate synthetase-like 2
|
|
chr2_-_51790163
|
55.215
|
NM_001141982
|
Rbm43
|
RNA binding motif protein 43
|
|
chr11_+_82988687
|
55.050
|
NM_011410
|
Slfn4
|
schlafen 4
|
|
chr17_-_34999458
|
54.777
|
NM_001142706
NM_008198
|
Cfb
|
complement factor B
|
|
chr6_+_122902755
|
54.540
|
|
Clec4a3
|
C-type lectin domain family 4, member a3
|
|
chr17_-_36179615
|
52.961
|
NM_010397
NM_010399
|
H2-T22
H2-T9
|
histocompatibility 2, T region locus 22
histocompatibility 2, T region locus 9
|
|
chr7_-_111613624
|
52.520
|
NM_009099
|
Trim30a
|
tripartite motif-containing 30A
|
|
chr7_-_134158053
|
52.404
|
NM_080638
|
Mvp
|
major vault protein
|
|
chr1_-_175529396
|
52.280
|
NM_001177349
NM_001177350
|
Pydc4
|
pyrin domain containing 4
|
|
chr3_+_127494295
|
52.203
|
|
Tifa
|
TRAF-interacting protein with forkhead-associated domain
|
|
chr1_-_172906094
|
50.910
|
NM_001077189
NM_010187
|
Fcgr2b
|
Fc receptor, IgG, low affinity IIb
|
|
chr13_-_101087625
|
50.684
|
NM_021545
|
Naip6
Naip7
|
NLR family, apoptosis inhibitory protein 6
NLR family, apoptosis inhibitory protein 7
|
|
chr1_-_186556046
|
50.638
|
|
Hlx
|
H2.0-like homeobox
|
|
chr11_+_81928683
|
50.105
|
NM_021443
|
Ccl8
|
chemokine (C-C motif) ligand 8
|
|
chr10_+_127707555
|
49.971
|
NM_019963
|
Stat2
|
signal transducer and activator of transcription 2
|
|
chr13_-_23802635
|
49.892
|
NM_010424
|
Hfe
|
hemochromatosis
|
|
chr17_-_35137278
|
49.390
|
|
D17H6S56E-5
|
DNA segment, Chr 17, human D6S56E 5
|
|
chr6_+_70676428
|
48.726
|
|
|
|
|
chr3_+_142223490
|
47.963
|
|
Gbp3
|
guanylate binding protein 3
|
|
chr8_+_70018744
|
47.768
|
NM_001168577
NM_010874
|
Nat2
|
N-acetyltransferase 2 (arylamine N-acetyltransferase)
|
|
chr11_+_58028478
|
46.311
|
NM_019440
|
Irgm2
|
immunity-related GTPase family M member 2
|
|
chr2_-_51828671
|
46.293
|
NM_001141948
NM_001141949
NM_019401
|
Nmi
|
N-myc (and STAT) interactor
|
|
chr7_-_152469796
|
46.233
|
NM_146206
|
Tpcn2
|
two pore segment channel 2
|
|
chr6_-_54922563
|
45.640
|
NM_172729
|
Nod1
|
nucleotide-binding oligomerization domain containing 1
|
|
chr1_-_186556329
|
45.227
|
NM_008250
|
Hlx
|
H2.0-like homeobox
|
|
chr19_+_12535536
|
44.775
|
|
Mpeg1
|
macrophage expressed gene 1
|
|
chr18_+_60535680
|
44.567
|
NM_001146275
NM_021792
|
Iigp1
|
interferon inducible GTPase 1
|
|
chr6_-_3349470
|
44.484
|
NM_010156
|
Samd9l
|
sterile alpha motif domain containing 9-like
|
|
chr12_+_33063902
|
44.389
|
|
2010109K11Rik
|
RIKEN cDNA 2010109K11 gene
|
|
chr7_-_111501738
|
43.620
|
NM_175677
NM_001146007
|
9230105E10Rik
|
RIKEN cDNA 9230105E10 gene
|
|
chr17_-_35137291
|
43.615
|
|
D17H6S56E-5
|
DNA segment, Chr 17, human D6S56E 5
|
|
chr13_+_74777303
|
43.578
|
NM_030711
|
Erap1
|
endoplasmic reticulum aminopeptidase 1
|
|
chr7_+_31206749
|
43.549
|
|
Nfkbid
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, delta
|
|
chr15_-_77363703
|
42.852
|
NM_175391
|
Apol7c
|
apolipoprotein L 7c
|
|
chr5_-_101148612
|
42.778
|
NM_152803
|
Hpse
|
heparanase
|
|
chr1_-_158604396
|
42.424
|
NM_023141
|
Tor3a
|
torsin family 3, member A
|
|
chr14_+_56197282
|
41.024
|
NM_011189
|
Psme1
|
proteasome (prosome, macropain) 28 subunit, alpha
|
|
chr17_+_18024787
|
40.909
|
NM_008039
|
Fpr2
Fpr3
|
formyl peptide receptor 2
formyl peptide receptor 3
|
|
chr7_-_148196606
|
40.723
|
|
Ifitm3
|
interferon induced transmembrane protein 3
|
|
chr7_-_148196526
|
40.576
|
|
Ifitm3
|
interferon induced transmembrane protein 3
|
|
chr7_-_148196635
|
40.559
|
NM_025378
|
Ifitm3
|
interferon induced transmembrane protein 3
|
|
chr4_-_86314560
|
40.419
|
|
Plin2
|
perilipin 2
|
|
chr13_+_33096472
|
40.412
|
|
Serpinb9
|
serine (or cysteine) peptidase inhibitor, clade B, member 9
|
|
chr9_-_119248925
|
39.872
|
NM_010851
|
Myd88
|
myeloid differentiation primary response gene 88
|
|
chr5_-_121262512
|
39.621
|
NM_033541
|
Oas1c
|
2'-5' oligoadenylate synthetase 1C
|
|
chr3_-_135271220
|
39.551
|
|
Nfkb1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 1, p105
|
|
chr2_-_126758750
|
39.230
|
|
2010106G01Rik
|
RIKEN cDNA 2010106G01 gene
|
|
chr6_+_127396847
|
39.041
|
|
Parp11
|
poly (ADP-ribose) polymerase family, member 11
|
|
chr1_+_177562307
|
39.023
|
|
Kmo
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase)
|
|
chr7_+_111392998
|
39.015
|
|
Trim34a
|
tripartite motif-containing 34A
|
|
chr18_-_67405109
|
38.917
|
NM_172630
|
Mppe1
|
metallophosphoesterase 1
|
|
chr14_-_59960166
|
38.798
|
NM_001164327
|
Gm4902
|
predicted gene 4902
|
|
chr9_-_106378901
|
38.473
|
NM_145619
|
Parp3
|
poly (ADP-ribose) polymerase family, member 3
|
|
chr2_+_164881126
|
38.322
|
NM_011611
NM_170702
NM_170703
NM_170704
|
Cd40
|
CD40 antigen
|
|
chr6_+_123212124
|
37.978
|
NM_001163161
NM_010819
|
Clec4d
|
C-type lectin domain family 4, member d
|