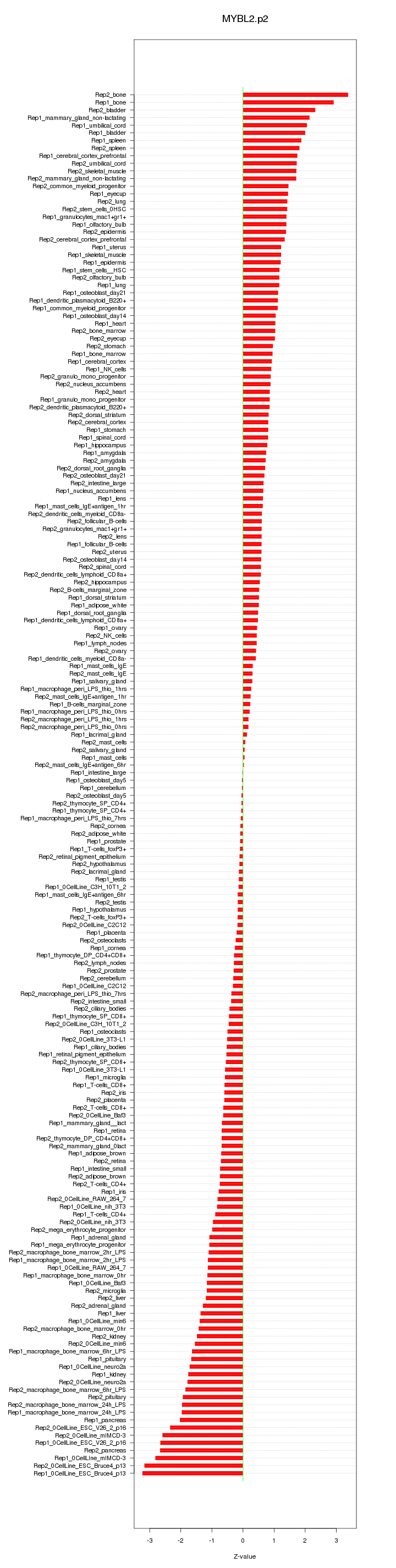
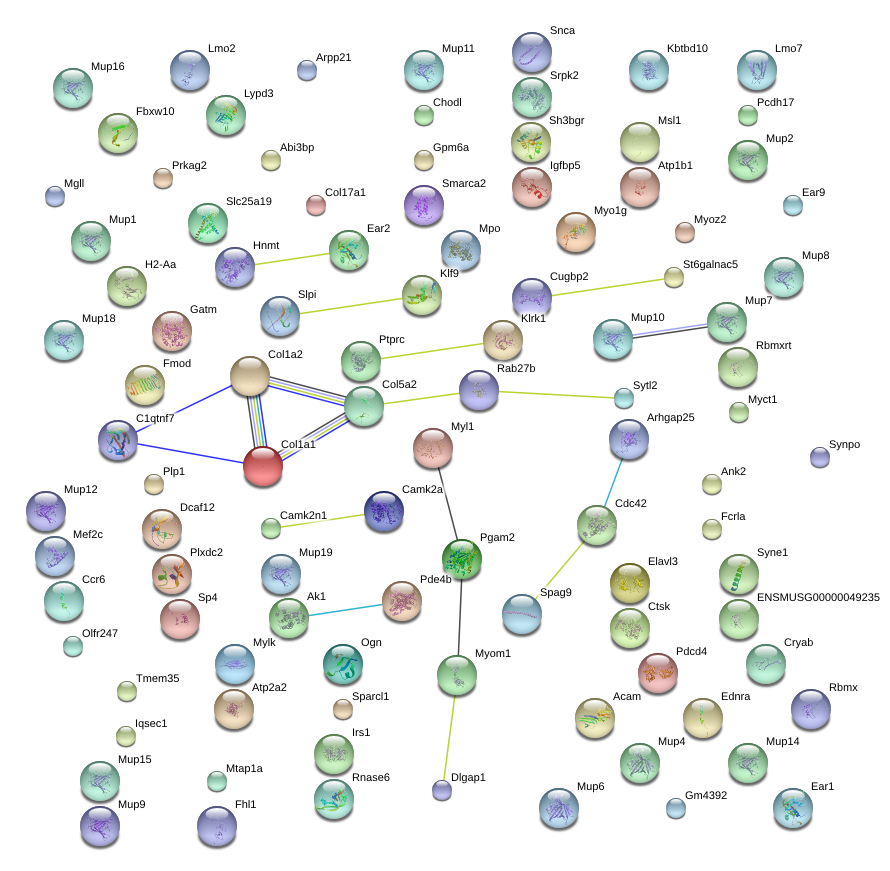
|
chr5_-_104543106
|
21.059
|
NM_010097
|
Sparcl1
|
SPARC-like 1
|
|
chr13_+_49704471
|
18.717
|
|
Ogn
|
osteoglycin
|
|
chr16_+_35002066
|
13.966
|
|
Mylk
|
myosin, light polypeptide kinase
|
|
chr11_+_93905443
|
12.478
|
|
Spag9
|
sperm associated antigen 9
|
|
chr6_+_70676428
|
12.473
|
|
|
|
|
chr13_+_83803830
|
11.751
|
|
Mef2c
|
myocyte enhancer factor 2C
|
|
chr11_+_87607531
|
11.364
|
|
Mpo
|
myeloperoxidase
|
|
chr16_+_96422243
|
11.089
|
|
Sh3bgr
|
SH3-binding domain glutamic acid-rich protein
|
|
chr11_+_87607285
|
10.710
|
NM_010824
|
Mpo
|
myeloperoxidase
|
|
chr11_+_93905557
|
10.568
|
|
Spag9
|
sperm associated antigen 9
|
|
chr4_-_60594496
|
10.201
|
|
Mup1
|
major urinary protein 1
|
|
chr4_-_60889249
|
9.959
|
NM_001134674
|
Mup13
Mup19
Mup1
|
major urinary protein 13
major urinary protein 19
major urinary protein 1
|
|
chr4_-_60594469
|
9.716
|
|
Mup1
|
major urinary protein 1
|
|
chrX_+_54032858
|
9.573
|
NM_001077362
|
Fhl1
|
four and a half LIM domains 1
|
|
chr2_+_103798150
|
9.494
|
NM_001142337
|
Lmo2
|
LIM domain only 2
|
|
chr4_+_138012860
|
9.261
|
|
Camk2n1
|
calcium/calmodulin-dependent protein kinase II inhibitor 1
|
|
chr16_+_96422076
|
9.201
|
NM_015825
|
Sh3bgr
|
SH3-binding domain glutamic acid-rich protein
|
|
chr16_+_35001358
|
8.902
|
|
Mylk
|
myosin, light polypeptide kinase
|
|
chr6_-_60779848
|
8.699
|
NM_009221
|
Snca
|
synuclein, alpha
|
|
chrX_+_54032914
|
8.674
|
|
Fhl1
|
four and a half LIM domains 1
|
|
chr16_+_34995000
|
8.438
|
|
Mylk
|
myosin, light polypeptide kinase
|
|
chr6_+_70676803
|
8.390
|
|
|
|
|
chr11_+_94811409
|
8.189
|
|
Col1a1
|
collagen, type I, alpha 1
|
|
chr11_+_93905501
|
7.863
|
NM_001025428
NM_001025429
NM_001025430
|
Spag9
|
sperm associated antigen 9
|
|
chr2_-_6466116
|
7.669
|
|
Celf2
|
CUGBP, Elav-like family member 2
|
|
chr18_-_60783889
|
7.563
|
|
Synpo
|
synaptopodin
|
|
chr1_-_82232392
|
7.481
|
|
Irs1
|
insulin receptor substrate 1
|
|
chr3_+_95303177
|
7.217
|
NM_007802
|
Ctsk
|
cathepsin K
|
|
chrX_+_54043764
|
7.130
|
|
Fhl1
|
four and a half LIM domains 1
|
|
chr6_+_4471231
|
7.103
|
|
Col1a2
|
collagen, type I, alpha 2
|
|
chr9_+_40590943
|
6.791
|
|
9030425E11Rik
|
RIKEN cDNA 9030425E11 gene
|
|
chr19_+_26822892
|
6.697
|
NM_026003
|
Smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2
|
|
chr7_+_97535490
|
6.524
|
NM_001040087
NM_001040088
|
Sytl2
|
synaptotagmin-like 2
|
|
chrX_+_133357268
|
6.483
|
NM_011123
|
Plp1
|
proteolipid protein (myelin) 1
|
|
chr16_+_96422285
|
6.292
|
|
Sh3bgr
|
SH3-binding domain glutamic acid-rich protein
|
|
chr2_-_164182009
|
6.267
|
|
Slpi
|
secretory leukocyte peptidase inhibitor
|
|
chr6_-_90611092
|
6.168
|
|
Iqsec1
|
IQ motif and Sec7 domain 1
|
|
chr7_+_97535516
|
5.945
|
|
Sytl2
|
synaptotagmin-like 2
|
|
chr1_-_72906185
|
5.917
|
|
Igfbp5
|
insulin-like growth factor binding protein 5
|
|
chr1_-_166368439
|
5.648
|
|
Atp1b1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide
|
|
chr4_-_60675161
|
5.546
|
NM_001164526
NM_001134644
NM_001045550
NM_008647
|
Mup10
Mup11
LOC100048885
Mup19
Mup1
Mup2
|
major urinary protein 10
major urinary protein 11
major urinary protein LOC100048885
major urinary protein 19
major urinary protein 1
major urinary protein 2
|
|
chr1_-_140071818
|
5.535
|
NM_001111316
NM_011210
|
Ptprc
|
protein tyrosine phosphatase, receptor type, C
|
|
chr16_+_56477956
|
5.453
|
NM_001014399
NM_001014422
NM_001014423
NM_001014424
NM_178790
|
Abi3bp
|
ABI gene family, member 3 (NESH) binding protein
|
|
chr10_+_4978047
|
5.395
|
|
Syne1
|
synaptic nuclear envelope 1
|
|
chr1_+_135934035
|
5.378
|
NM_021355
|
Fmod
|
fibromodulin
|
|
chr3_-_126646303
|
5.321
|
NM_001034168
|
Ank2
|
ankyrin 2, brain
|
|
chr2_+_69508174
|
5.075
|
NM_001081087
|
Kbtbd10
|
kelch repeat and BTB (POZ) domain containing 10
|
|
chr9_+_50561121
|
5.056
|
|
Cryab
|
crystallin, alpha B
|
|
chr14_-_104213981
|
5.025
|
|
|
|
|
chr9_+_50561092
|
5.014
|
|
Cryab
|
crystallin, alpha B
|
|
chr4_+_102260162
|
4.963
|
NM_001177982
|
Pde4b
|
phosphodiesterase 4B, cAMP specific
|
|
chr18_-_70301202
|
4.916
|
NM_001082553
|
Rab27b
|
RAB27b, member RAS oncogene family
|
|
chr1_-_45560001
|
4.795
|
NM_007737
|
Col5a2
|
collagen, type V, alpha 2
|
|
chr4_-_136891931
|
4.731
|
|
Cdc42
|
cell division cycle 42 homolog (S. cerevisiae)
|
|
chr9_+_50560862
|
4.664
|
NM_009964
|
Cryab
|
crystallin, alpha B
|
|
chr19_+_54003803
|
4.637
|
|
Pdcd4
|
programmed cell death 4
|
|
chr11_-_5703752
|
4.603
|
NM_018870
|
Pgam2
|
phosphoglycerate mutase 2
|
|
chr1_-_66981900
|
4.575
|
NM_001113387
|
Myl1
|
myosin, light polypeptide 1
|
|
chrX_+_133371611
|
4.466
|
|
Plp1
|
proteolipid protein (myelin) 1
|
|
chr1_-_172857670
|
4.372
|
NM_001160215
NM_145141
|
Fcrla
|
Fc receptor-like A
|
|
chr11_-_5703677
|
4.299
|
|
Pgam2
|
phosphoglycerate mutase 2
|
|
chr19_+_23215725
|
4.269
|
|
Klf9
|
Kruppel-like factor 9
|
|
chr3_-_122737855
|
4.202
|
NM_021503
|
Myoz2
|
myozenin 2
|
|
chrX_-_54646073
|
4.136
|
NM_001166623
NM_011252
|
Rbmx
|
RNA binding motif protein, X chromosome
|
|
chr18_+_61085283
|
3.966
|
NM_177407
|
Camk2a
|
calcium/calmodulin-dependent protein kinase II alpha
|
|
chr2_+_32484995
|
3.965
|
NM_021515
|
Ak1
|
adenylate kinase 1
|
|
chr4_-_60594959
|
3.944
|
NM_001122647
|
Mup10
LOC100048884
Mup1
|
major urinary protein 10
novel member of the major urinary protein (Mup) gene family
major urinary protein 1
|
|
chr14_+_84933363
|
3.831
|
|
Pcdh17
|
protocadherin 17
|
|
chr19_+_23241546
|
3.823
|
|
Klf9
|
Kruppel-like factor 9
|
|
chr9_+_50561381
|
3.807
|
|
Cryab
|
crystallin, alpha B
|
|
chr4_+_102262447
|
3.764
|
NM_001177981
|
Pde4b
|
phosphodiesterase 4B, cAMP specific
|
|
chr17_+_8437922
|
3.749
|
NM_001190338
|
Ccr6
|
chemokine (C-C motif) receptor 6
|
|
chr1_-_140071630
|
3.717
|
|
Ptprc
|
protein tyrosine phosphatase, receptor type, C
|
|
chr17_+_70871444
|
3.601
|
NM_001128180
|
Dlgap1
|
discs, large (Drosophila) homolog-associated protein 1
|
|
chr10_-_4752710
|
3.596
|
NM_026793
|
Myct1
|
myc target 1
|
|
chr18_+_61085303
|
3.391
|
|
Camk2a
|
calcium/calmodulin-dependent protein kinase II alpha
|
|
chr2_+_16277963
|
3.372
|
|
Plxdc2
|
plexin domain containing 2
|
|
chr2_-_122420914
|
3.363
|
|
Gatm
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase)
|
|
chr12_-_119474010
|
3.338
|
|
Sp4
|
trans-acting transcription factor 4
|
|
chr5_-_23012407
|
3.296
|
|
Srpk2
|
serine/arginine-rich protein specific kinase 2
|
|
chr5_-_24606229
|
3.261
|
|
Prkag2
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit
|
|
chr2_-_23904343
|
3.220
|
|
Hnmt
|
histamine N-methyltransferase
|
|
chr6_-_87483228
|
3.144
|
NM_001037727
|
Arhgap25
|
Rho GTPase activating protein 25
|
|
chr11_+_98667577
|
3.116
|
|
Msl1
|
male-specific lethal 1 homolog (Drosophila)
|
|
chr4_-_61179982
|
3.024
|
|
Mup1
|
major urinary protein 1
|
|
chr2_-_164182242
|
3.018
|
NM_011414
|
Slpi
|
secretory leukocyte peptidase inhibitor
|
|
chr11_+_62633754
|
2.935
|
NM_053169
|
Trim16
|
tripartite motif-containing 16
|
|
chr14_-_44501032
|
2.894
|
NM_053112
|
Ear10
|
eosinophil-associated, ribonuclease A family, member 10
|
|
chr9_+_50561315
|
2.887
|
|
Cryab
|
crystallin, alpha B
|
|
chr4_-_60736086
|
2.886
|
|
Mup10
Mup1
|
major urinary protein 10
major urinary protein 1
|
|
chr19_-_47766512
|
2.840
|
NM_007732
|
Col17a1
|
collagen, type XVII, alpha 1
|
|
chr8_-_80248234
|
2.828
|
|
Ednra
|
endothelin receptor type A
|
|
chr5_+_43906796
|
2.803
|
NM_001135172
|
C1qtnf7
|
C1q and tumor necrosis factor related protein 7
|
|
chr5_+_43907004
|
2.758
|
|
C1qtnf7
|
C1q and tumor necrosis factor related protein 7
|
|
chr17_+_71368860
|
2.728
|
NM_001083934
NM_010867
|
Myom1
|
myomesin 1
|
|
chr5_-_122906470
|
2.683
|
|
Atp2a2
|
ATPase, Ca++ transporting, cardiac muscle, slow twitch 2
|
|
chr19_+_23215704
|
2.671
|
NM_010638
|
Klf9
|
Kruppel-like factor 9
|
|
chr2_+_121121189
|
2.656
|
NM_001173506
|
Mtap1a
|
microtubule-associated protein 1 A
|
|
chr6_-_129572590
|
2.629
|
NM_033078
|
Klrk1
|
killer cell lectin-like receptor subfamily K, member 1
|
|
chr17_-_34424623
|
2.570
|
NM_010378
|
H2-Aa
|
histocompatibility 2, class II antigen A, alpha
|
|
chr5_-_144266956
|
2.507
|
|
|
|
|
chr9_-_21820860
|
2.486
|
|
Elavl3
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 3 (Hu antigen C)
|
|
chr3_-_152645206
|
2.471
|
|
St6galnac5
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 5
|
|
chr9_-_112090150
|
2.449
|
NM_033264
NM_001177615
NM_001177620
NM_028755
|
Arpp21
|
cyclic AMP-regulated phosphoprotein, 21
|
|
chr4_-_41261743
|
2.445
|
|
Dcaf12
|
DDB1 and CUL4 associated factor 12
|
|
chr7_+_25421551
|
2.441
|
|
Lypd3
|
Ly6/Plaur domain containing 3
|
|
chr6_+_88674821
|
2.427
|
|
Mgll
|
monoglyceride lipase
|
|
chr4_-_41261815
|
2.413
|
|
Dcaf12
|
DDB1 and CUL4 associated factor 12
|
|
chr14_+_102239673
|
2.393
|
|
Lmo7
|
LIM domain only 7
|
|
chr16_+_78931190
|
2.350
|
NM_139134
|
Chodl
|
chondrolectin
|
|
chrX_+_130829929
|
2.332
|
|
Tmem35
|
transmembrane protein 35
|
|
chr11_-_6420931
|
2.329
|
NM_178440
|
Myo1g
|
myosin IG
|
|
chr14_+_51748742
|
2.323
|
NM_030098
|
Rnase6
|
ribonuclease, RNase A family, 6
|
|
chr19_+_12040311
|
2.314
|
NM_001033174
|
Osbp
|
oxysterol binding protein
|
|
chr2_+_125860507
|
2.305
|
|
Fgf7
|
fibroblast growth factor 7
|
|
chr11_-_30000031
|
2.300
|
|
Spnb2
|
spectrin beta 2
|
|
chr6_-_119146426
|
2.297
|
NM_009781
|
Cacna1c
|
calcium channel, voltage-dependent, L type, alpha 1C subunit
|
|
chr14_+_124059330
|
2.273
|
NM_145467
|
Itgbl1
|
integrin, beta-like 1
|
|
chr1_+_89967340
|
2.236
|
NM_201644
|
Ugt1a9
|
UDP glucuronosyltransferase 1 family, polypeptide A9
|
|
chr6_-_54543167
|
2.235
|
|
Fkbp14
|
FK506 binding protein 14
|
|
chr4_+_102242664
|
2.219
|
NM_001177980
|
Pde4b
|
phosphodiesterase 4B, cAMP specific
|
|
chr11_-_21222127
|
2.218
|
|
Ugp2
|
UDP-glucose pyrophosphorylase 2
|
|
chr5_-_24501570
|
2.216
|
|
Prkag2
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit
|
|
chr4_+_94406190
|
2.209
|
|
Tek
|
endothelial-specific receptor tyrosine kinase
|
|
chr8_+_72758454
|
2.208
|
|
Sugp2
|
SURP and G patch domain containing 2
|
|
chr1_-_194681914
|
2.204
|
NM_198247
|
Sertad4
|
SERTA domain containing 4
|
|
chr7_-_110962416
|
2.197
|
NM_008220
NM_016956
|
Hbb-b1
Hbb-b2
|
hemoglobin, beta adult major chain
hemoglobin, beta adult minor chain
|
|
chr2_-_30141815
|
2.164
|
|
Dolk
|
dolichol kinase
|
|
chr11_-_6420904
|
2.160
|
|
Myo1g
|
myosin IG
|
|
chr9_-_70452528
|
2.145
|
|
Fam63b
|
family with sequence similarity 63, member B
|
|
chr8_+_46593141
|
2.096
|
NM_172752
|
Sorbs2
|
sorbin and SH3 domain containing 2
|
|
chr5_+_104631633
|
2.090
|
NM_016779
|
Dmp1
|
dentin matrix protein 1
|
|
chr9_-_16182674
|
2.086
|
NM_001080814
|
Fat3
|
FAT tumor suppressor homolog 3 (Drosophila)
|
|
chr7_+_25421586
|
2.054
|
NM_133743
|
Lypd3
|
Ly6/Plaur domain containing 3
|
|
chr14_+_52473672
|
2.050
|
NM_017385
|
Ear7
|
eosinophil-associated, ribonuclease A family, member 7
|
|
chr16_+_33794452
|
2.034
|
|
Muc13
|
mucin 13, epithelial transmembrane
|
|
chr16_+_45093784
|
2.022
|
|
Ccdc80
|
coiled-coil domain containing 80
|
|
chr6_-_54543121
|
2.014
|
NM_153573
|
Fkbp14
|
FK506 binding protein 14
|
|
chr16_+_34994935
|
2.005
|
|
Mylk
|
myosin, light polypeptide kinase
|
|
chr8_+_72758086
|
1.995
|
NM_001168290
NM_172755
|
Sugp2
|
SURP and G patch domain containing 2
|
|
chr2_+_14310024
|
1.986
|
NM_001012305
|
Slc39a12
|
solute carrier family 39 (zinc transporter), member 12
|
|
chrX_+_130829763
|
1.984
|
NM_026239
|
Tmem35
|
transmembrane protein 35
|
|
chr6_+_68375560
|
1.981
|
|
Igkv16-104
|
immunoglobulin kappa variable 16-104
|
|
chr9_+_14080744
|
1.959
|
NM_030261
|
Sesn3
|
sestrin 3
|
|
chr9_+_75623153
|
1.936
|
NM_007555
|
Bmp5
|
bone morphogenetic protein 5
|
|
chr16_+_43619057
|
1.935
|
|
Zbtb20
|
zinc finger and BTB domain containing 20
|
|
chr9_-_79566271
|
1.931
|
NM_007730
|
Col12a1
|
collagen, type XII, alpha 1
|
|
chr1_+_34356614
|
1.930
|
|
Dst
|
dystonin
|
|
chr10_-_41905549
|
1.930
|
|
Foxo3
|
forkhead box O3
|
|
chr3_-_146433544
|
1.892
|
NM_001164199
|
Prkacb
|
protein kinase, cAMP dependent, catalytic, beta
|
|
chr3_-_92289795
|
1.851
|
NM_009264
|
Sprr1a
|
small proline-rich protein 1A
|
|
chr8_+_56040553
|
1.848
|
|
Gpm6a
|
glycoprotein m6a
|
|
chr9_+_44448105
|
1.842
|
|
Ddx6
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 6
|
|
chr1_+_90003410
|
1.840
|
|
Ugt1a6b
|
UDP glucuronosyltransferase 1 family, polypeptide A6B
|
|
chr5_+_111846284
|
1.835
|
|
Mn1
|
meningioma 1
|
|
chr6_+_124254668
|
1.831
|
NM_001170395
NM_053094
|
Cd163
|
CD163 antigen
|
|
chrX_+_146981959
|
1.812
|
NM_001102446
NM_009653
|
Alas2
|
aminolevulinic acid synthase 2, erythroid
|
|
chr3_+_136566015
|
1.798
|
|
Ppp3ca
|
protein phosphatase 3, catalytic subunit, alpha isoform
|
|
chr19_-_21547035
|
1.792
|
|
Gda
|
guanine deaminase
|
|
chr12_+_105452740
|
1.745
|
NM_009251
NM_001168294
NM_001168295
NM_001033335
|
Serpina3g
Serpina3f
Serpina3h
|
serine (or cysteine) peptidase inhibitor, clade A, member 3G
serine (or cysteine) peptidase inhibitor, clade A, member 3F
serine (or cysteine) peptidase inhibitor, clade A, member 3H
|
|
chr11_+_115168476
|
1.740
|
NM_172801
|
Otop2
|
otopetrin 2
|
|
chr18_+_69753146
|
1.737
|
|
Tcf4
|
transcription factor 4
|
|
chr2_+_55288297
|
1.737
|
|
Kcnj3
|
potassium inwardly-rectifying channel, subfamily J, member 3
|
|
chr3_-_92460819
|
1.732
|
NM_026822
|
Lce1b
|
late cornified envelope 1B
|
|
chr6_+_96063137
|
1.717
|
NM_182808
|
Fam19a1
|
family with sequence similarity 19, member A1
|
|
chr19_-_21547072
|
1.713
|
NM_010266
|
Gda
|
guanine deaminase
|
|
chr17_+_75864877
|
1.711
|
NM_207246
|
Rasgrp3
|
RAS, guanyl releasing protein 3
|
|
chr8_-_72425538
|
1.707
|
|
Ndufa13
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 13
|
|
chr19_-_23522811
|
1.676
|
NM_174857
|
Mamdc2
|
MAM domain containing 2
|
|
chr16_+_33794122
|
1.674
|
NM_010739
|
Muc13
|
mucin 13, epithelial transmembrane
|
|
chr12_+_53800369
|
1.674
|
NM_198111
|
Akap6
|
A kinase (PRKA) anchor protein 6
|
|
chr11_-_118163704
|
1.669
|
|
Timp2
|
tissue inhibitor of metalloproteinase 2
|
|
chr12_+_72149332
|
1.648
|
|
Arid4a
|
AT rich interactive domain 4A (RBP1-like)
|
|
chr14_-_122138082
|
1.604
|
NM_001128307
NM_001128308
|
Dock9
|
dedicator of cytokinesis 9
|
|
chr15_-_78034565
|
1.598
|
NM_013645
|
Pvalb
|
parvalbumin
|
|
chr11_-_30839546
|
1.596
|
|
Erlec1
|
endoplasmic reticulum lectin 1
|
|
chr18_-_39078304
|
1.595
|
NM_010197
|
Fgf1
|
fibroblast growth factor 1
|
|
chr16_-_14291427
|
1.589
|
NM_001161775
NM_013607
|
Myh11
|
myosin, heavy polypeptide 11, smooth muscle
|
|
chr4_-_41261874
|
1.571
|
|
Dcaf12
|
DDB1 and CUL4 associated factor 12
|
|
chr3_-_152645143
|
1.570
|
NM_012028
|
St6galnac5
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 5
|
|
chr8_-_37631532
|
1.559
|
|
Dlc1
|
deleted in liver cancer 1
|
|
chr17_+_35373506
|
1.556
|
NM_023179
|
Atp6v1g2
|
ATPase, H+ transporting, lysosomal V1 subunit G2
|
|
chr2_+_125860377
|
1.543
|
NM_008008
|
Fgf7
|
fibroblast growth factor 7
|
|
chr11_+_94812919
|
1.539
|
|
Col1a1
|
collagen, type I, alpha 1
|
|
chr14_+_45607785
|
1.538
|
NM_008964
|
Ptger2
|
prostaglandin E receptor 2 (subtype EP2)
|
|
chr3_+_7366586
|
1.536
|
NM_008862
|
Pkia
|
protein kinase inhibitor, alpha
|
|
chr1_-_45460310
|
1.534
|
|
Col5a2
|
collagen, type V, alpha 2
|
|
chr15_-_91022198
|
1.527
|
NM_011994
|
Abcd2
|
ATP-binding cassette, sub-family D (ALD), member 2
|
|
chr5_+_65522325
|
1.512
|
NM_175174
|
Klhl5
|
kelch-like 5 (Drosophila)
|
|
chr13_-_36826104
|
1.505
|
|
Nrn1
|
neuritin 1
|
|
chrX_-_73088288
|
1.501
|
NM_001166454
|
Pls3
|
plastin 3 (T-isoform)
|
|
chr10_-_118293979
|
1.492
|
|
Dyrk2
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 2
|
|
chr5_+_90889937
|
1.486
|
|
Alb
|
albumin
|
|
chr19_-_46401618
|
1.485
|
NM_028627
|
Psd
|
pleckstrin and Sec7 domain containing
|
|
chr4_-_137869546
|
1.479
|
|
Pink1
|
PTEN induced putative kinase 1
|
|
chr8_-_42127124
|
1.468
|
|
Mtus1
|
mitochondrial tumor suppressor 1
|
|
chr7_-_107805466
|
1.456
|
NM_001163186
NM_001163187
|
Plekhb1
|
pleckstrin homology domain containing, family B (evectins) member 1
|
|
chr15_-_91022165
|
1.452
|
|
Abcd2
|
ATP-binding cassette, sub-family D (ALD), member 2
|
|
chr2_+_16277924
|
1.451
|
NM_026162
|
Plxdc2
|
plexin domain containing 2
|
|
chr7_-_148615247
|
1.444
|
NM_021316
|
Cend1
|
cell cycle exit and neuronal differentiation 1
|