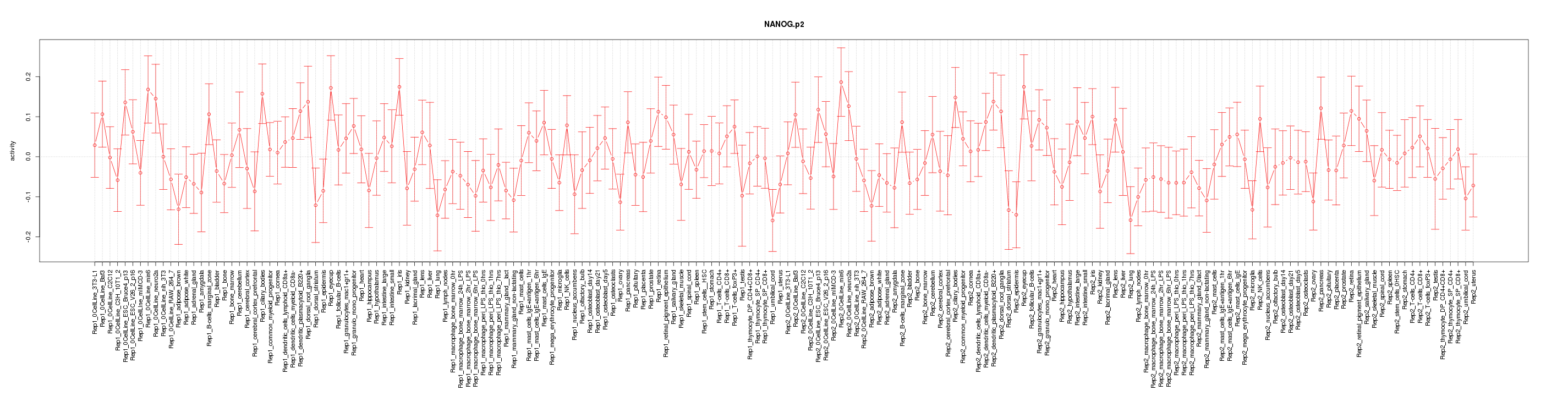
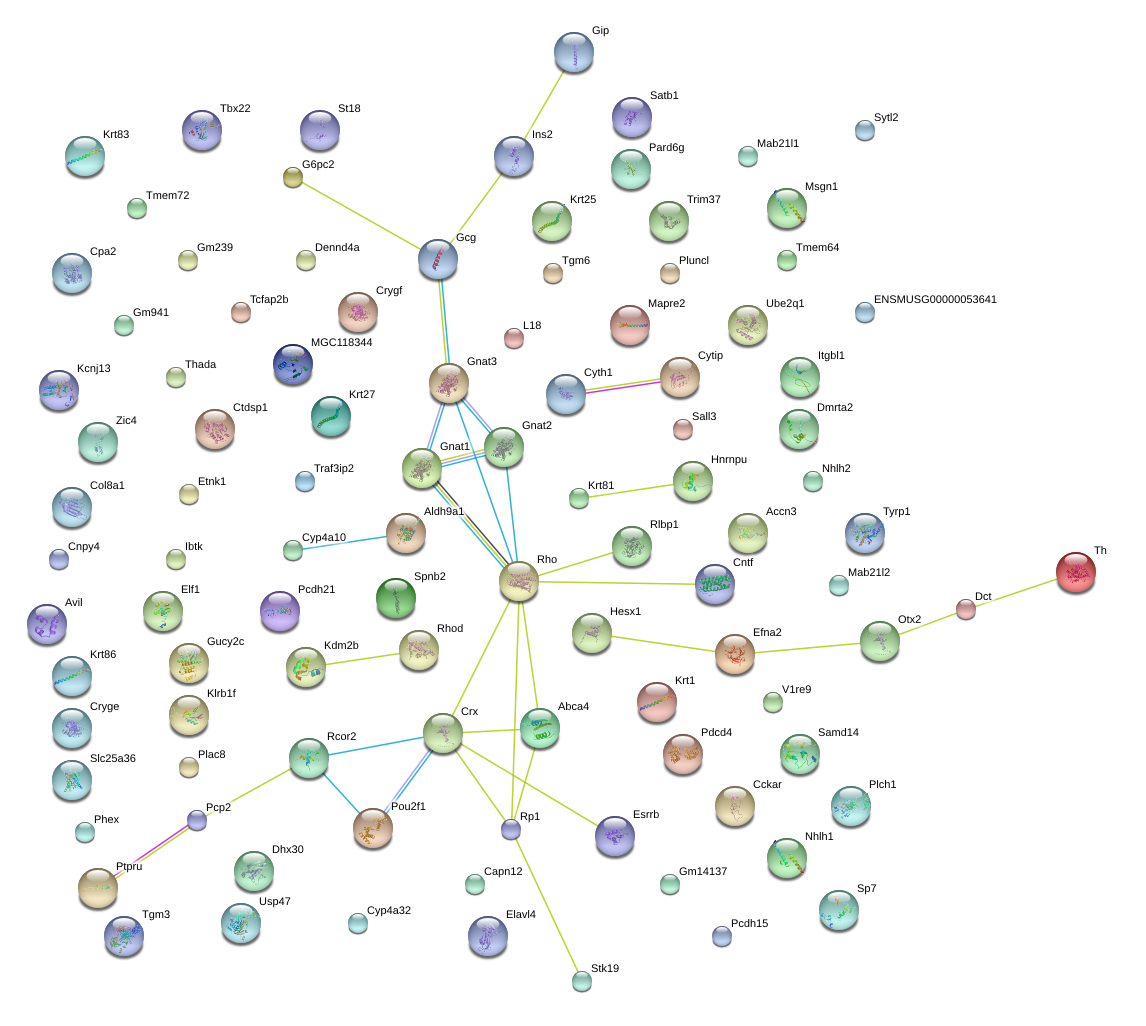
|
chr14_-_118450875
|
15.558
|
NM_010024
|
Dct
|
dopachrome tautomerase
|
|
chr4_+_80480116
|
13.078
|
NM_031202
|
Tyrp1
|
tyrosinase-related protein 1
|
|
chr14_-_49282546
|
11.901
|
NM_144841
|
Otx2
|
orthodenticle homolog 2 (Drosophila)
|
|
chr14_-_37911383
|
10.214
|
NM_130878
|
Cdhr1
|
cadherin-related family member 1
|
|
chr3_+_17695743
|
8.589
|
|
6430547I21Rik
|
RIKEN cDNA 6430547I21 gene
|
|
chr3_+_107895985
|
8.162
|
NM_008141
|
Gnat2
|
guanine nucleotide binding protein, alpha transducing 2
|
|
chr9_-_96978102
|
6.951
|
|
Slc25a36
|
solute carrier family 25, member 36
|
|
chr12_-_11215753
|
6.816
|
NM_019544
|
Msgn1
|
mesogenin 1
|
|
chr3_-_86352170
|
6.771
|
NM_011839
|
Mab21l2
|
mab-21-like 2 (C. elegans)
|
|
chr19_+_54003803
|
6.757
|
|
Pdcd4
|
programmed cell death 4
|
|
chr2_+_119000244
|
6.682
|
NM_001039223
|
Gm14137
|
predicted gene 14137
|
|
chr19_-_12840121
|
6.652
|
NM_170786
|
Cntf
|
ciliary neurotrophic factor
|
|
chr10_+_73284612
|
6.585
|
NM_001142735
NM_001142736
NM_001142737
NM_001142738
NM_001142739
NM_001142740
NM_001142741
NM_001142742
NM_001142743
NM_001142746
NM_001142747
NM_001142748
NM_001142760
NM_023115
|
Pcdh15
|
protocadherin 15
|
|
chr3_+_121747377
|
6.566
|
NM_007378
|
Abca4
|
ATP-binding cassette, sub-family A (ABC1), member 4
|
|
chr1_+_6720153
|
6.284
|
|
St18
|
suppression of tumorigenicity 18
|
|
chr14_+_79915429
|
6.232
|
|
Elf1
|
E74-like factor 1
|
|
chr4_-_109959366
|
6.179
|
NM_001163399
NM_010488
|
Elavl4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr1_-_4350323
|
6.031
|
NM_011283
|
Rp1
|
retinitis pigmentosa 1 (human)
|
|
chr2_-_62321694
|
5.899
|
NM_008100
|
Gcg
|
glucagon
|
|
chr11_-_99184254
|
5.689
|
NM_133730
|
Krt25
|
keratin 25
|
|
chr17_-_51878720
|
5.660
|
|
Satb1
|
special AT-rich sequence binding protein 1
|
|
chr2_+_129838084
|
5.551
|
NM_009374
|
Tgm3
|
transglutaminase 3, E polypeptide
|
|
chr3_+_101814059
|
5.465
|
NM_178777
|
Nhlh2
|
nescient helix loop helix 2
|
|
chr9_+_64658817
|
5.323
|
NM_001162917
|
Dennd4a
|
DENN/MADD domain containing 4A
|
|
chr6_-_72382855
|
5.252
|
|
|
|
|
chr9_+_46721685
|
5.109
|
|
2900052N01Rik
|
RIKEN cDNA 2900052N01 gene
|
|
chr13_-_64599099
|
4.864
|
NM_001195435
|
1190003K10Rik
|
RIKEN cDNA 1190003K10 gene
|
|
chr18_-_81183260
|
4.852
|
NM_178280
|
Sall3
|
sal-like 3 (Drosophila)
|
|
chr4_-_109959185
|
4.782
|
|
Elavl4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr11_+_87032504
|
4.745
|
|
Trim37
|
tripartite motif-containing 37
|
|
chr11_-_99212407
|
4.661
|
NM_010666
|
Krt27
|
keratin 27
|
|
chr7_-_150085870
|
4.488
|
NM_009377
|
Th
|
tyrosine hydroxylase
|
|
chr1_-_65097587
|
4.479
|
NM_007777
|
Cryge
|
crystallin, gamma E
|
|
chr6_-_136730254
|
4.357
|
NM_001127318
NM_145067
|
Gucy2c
|
guanylate cyclase 2c
|
|
chr7_+_97535516
|
4.334
|
|
Sytl2
|
synaptotagmin-like 2
|
|
chr2_+_69058005
|
4.274
|
NM_021331
|
G6pc2
|
glucose-6-phosphatase, catalytic, 2
|
|
chr2_+_153988342
|
4.235
|
NM_025990
|
2310021H06Rik
|
RIKEN cDNA 2310021H06 gene
|
|
chr6_+_128995918
|
4.207
|
NM_153094
|
Klrb1f
|
killer cell lectin-like receptor subfamily B member 1F
|
|
chr6_-_116666759
|
4.082
|
NM_178768
|
Tmem72
|
transmembrane protein 72
|
|
chr7_-_16465217
|
4.081
|
NM_007770
|
Crx
|
cone-rod homeobox containing gene
|
|
chr10_+_39332739
|
4.059
|
NM_134000
|
Traf3ip2
|
TRAF3 interacting protein 2
|
|
chr10_+_39332934
|
3.967
|
|
Traf3ip2
|
TRAF3 interacting protein 2
|
|
chr10_+_126437764
|
3.916
|
NM_009635
|
Avil
|
advillin
|
|
chr4_-_109078858
|
3.893
|
|
|
|
|
chr5_-_101001191
|
3.783
|
|
Plac8
|
placenta-specific 8
|
|
chrX_+_104871850
|
3.748
|
NM_145224
|
Tbx22
|
T-box 22
|
|
chr7_+_97535490
|
3.741
|
NM_001040087
NM_001040088
|
Sytl2
|
synaptotagmin-like 2
|
|
chr5_+_138628733
|
3.738
|
NM_178612
|
Cnpy4
|
canopy 4 homolog (zebrafish)
|
|
chr2_-_58012447
|
3.563
|
NM_139200
|
Cytip
|
cytohesin 1 interacting protein
|
|
chr9_-_85637765
|
3.555
|
NM_001081282
|
Ibtk
|
inhibitor of Bruton agammaglobulinemia tyrosine kinase
|
|
chr16_-_57754816
|
3.550
|
NM_007739
|
Col8a1
|
collagen, type VIII, alpha 1
|
|
chr15_+_101303908
|
3.536
|
NM_010667
|
Krt86
|
keratin 86
|
|
chr9_+_91263809
|
3.528
|
NM_009576
|
Zic4
|
zinc finger protein of the cerebellum 4
|
|
chr19_+_7344253
|
3.505
|
NM_054048
|
Rcor2
|
REST corepressor 2
|
|
chr1_+_19198994
|
3.420
|
NM_009334
|
Tcfap2b
|
transcription factor AP-2 beta
|
|
chr9_-_109986879
|
3.411
|
|
Dhx30
|
DEAH (Asp-Glu-Ala-His) box polypeptide 30
|
|
chr14_+_27813547
|
3.396
|
NM_010420
|
Hesx1
|
homeobox gene expressed in ES cells
|
|
chr1_+_169298077
|
3.394
|
|
Aldh9a1
|
aldehyde dehydrogenase 9, subfamily A1
|
|
chr7_-_26506741
|
3.170
|
|
|
|
|
chr12_+_72456427
|
3.128
|
|
|
|
|
chr7_-_86531587
|
3.093
|
|
Rlbp1
|
retinaldehyde binding protein 1
|
|
chr4_+_15211137
|
2.944
|
|
Tmem64
|
transmembrane protein 64
|
|
chr6_+_143156520
|
2.897
|
|
Etnk1
|
ethanolamine kinase 1
|
|
chr7_+_29666674
|
2.885
|
NM_001110807
|
Capn12
|
calpain 12
|
|
chr7_+_119253816
|
2.827
|
|
Usp47
|
ubiquitin specific peptidase 47
|
|
chr5_-_101001209
|
2.824
|
NM_139198
|
Plac8
|
placenta-specific 8
|
|
chr5_+_23919268
|
2.778
|
NM_183000
|
Accn3
|
amiloride-sensitive cation channel 3
|
|
chr9_+_92502076
|
2.776
|
|
|
|
|
chrX_-_153853209
|
2.771
|
NM_011077
|
Phex
|
phosphate regulating gene with homologies to endopeptidases on the X chromosome (hypophosphatemia, vitamin D resistant rickets)
|
|
chr2_+_129949020
|
2.748
|
NM_177726
|
Tgm6
|
transglutaminase 6
|
|
chr4_+_109650629
|
2.731
|
NM_172296
|
Dmrta2
|
doublesex and mab-3 related transcription factor like family A2
|
|
chr11_+_94871620
|
2.731
|
|
Samd14
|
sterile alpha motif domain containing 14
|
|
chr4_-_109960078
|
2.707
|
NM_001038698
|
Elavl4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr5_-_54098762
|
2.692
|
NM_009827
|
Cckar
|
cholecystokinin A receptor
|
|
chr14_+_124059330
|
2.685
|
NM_145467
|
Itgbl1
|
integrin, beta-like 1
|
|
chr12_-_88579550
|
2.670
|
NM_027744
|
4933437F05Rik
|
RIKEN cDNA 4933437F05 gene
|
|
chr3_+_89586320
|
2.668
|
|
Ube2q1
|
ubiquitin-conjugating enzyme E2Q (putative) 1
|
|
chr17_+_80706746
|
2.648
|
NM_001145452
|
Arhgef33
|
Rho guanine nucleotide exchange factor (GEF) 33
|
|
chr15_-_102196701
|
2.615
|
NM_130458
|
Sp7
|
Sp7 transcription factor 7
|
|
chr8_-_3625251
|
2.501
|
NM_008790
|
Pcp2
|
Purkinje cell protein 2 (L7)
|
|
chr11_+_95885858
|
2.479
|
NM_008119
|
Gip
|
gastric inhibitory polypeptide
|
|
chr1_-_180259089
|
2.473
|
|
Hnrnpu
|
heterogeneous nuclear ribonucleoprotein U
|
|
chr6_+_30491641
|
2.461
|
NM_001024698
|
Cpa2
|
carboxypeptidase A2, pancreatic
|
|
chr10_-_116333934
|
2.460
|
NM_001033333
|
Gm239
|
predicted gene 239
|
|
chr1_+_6720087
|
2.421
|
NM_173868
|
St18
|
suppression of tumorigenicity 18
|
|
chr1_-_89291303
|
2.396
|
NM_001110227
|
Kcnj13
|
potassium inwardly-rectifying channel, subfamily J, member 13
|
|
chr1_-_167865030
|
2.373
|
NM_198933
NM_198934
|
Pou2f1
|
POU domain, class 2, transcription factor 1
|
|
chr6_+_115881925
|
2.354
|
NM_145383
|
Rho
|
rhodopsin
|
|
chr16_+_88829238
|
2.336
|
NM_013713
|
Krtap15
|
keratin associated protein 15
|
|
chr5_-_123438517
|
2.320
|
NM_001005866
|
Kdm2b
|
lysine (K)-specific demethylase 2B
|
|
chr3_-_63654912
|
2.319
|
NM_001177732
NM_001177733
NM_183191
|
Plch1
|
phospholipase C, eta 1
|
|
chr12_+_87702066
|
2.257
|
NM_001159500
|
Esrrb
|
estrogen related receptor, beta
|
|
chr7_-_149865451
|
2.226
|
NM_001185083
NM_001185084
NM_008387
|
Ins2
|
insulin II
|
|
chrX_-_35087783
|
2.209
|
NM_001100465
|
Rhox2h
|
reproductive homeobox 2H
|
|
chr11_-_33103458
|
2.197
|
NM_019916
|
Tlx3
|
T-cell leukemia, homeobox 3
|
|
chr1_+_106665395
|
2.179
|
NM_011800
|
Cdh20
|
cadherin 20
|
|
chr11_+_11014018
|
2.170
|
NM_177033
|
Vwc2
|
von Willebrand factor C domain containing 2
|
|
chr1_-_79436564
|
2.132
|
NM_009129
|
Scg2
|
secretogranin II
|
|
chr19_-_29409041
|
2.132
|
NM_011272
|
Rln1
|
relaxin 1
|
|
chrX_+_34944974
|
2.127
|
NM_001085348
|
Rhox2e
|
reproductive homeobox 2E
|
|
chr11_-_94154354
|
2.127
|
|
Luc7l3
|
LUC7-like 3 (S. cerevisiae)
|
|
chr11_-_99427891
|
2.120
|
NM_023511
|
Krtap3-1
|
keratin associated protein 3-1
|
|
chr12_+_39510297
|
2.095
|
NM_001163154
|
Etv1
|
ets variant gene 1
|
|
chr7_-_91763744
|
2.054
|
|
Zfand6
|
zinc finger, AN1-type domain 6
|
|
chr3_-_154718282
|
2.035
|
NM_177066
|
Tnni3k
|
TNNI3 interacting kinase
|
|
chr15_-_101723333
|
2.025
|
NM_001033177
|
Krt76
|
keratin 76
|
|
chr10_-_20880559
|
2.019
|
NM_001198914
NM_010848
|
Myb
|
myeloblastosis oncogene
|
|
chr11_-_102807774
|
2.007
|
NM_011795
|
C1ql1
|
complement component 1, q subcomponent-like 1
|
|
chr9_+_69855789
|
1.996
|
|
Bnip2
|
BCL2/adenovirus E1B interacting protein 2
|
|
chr12_-_105711324
|
1.989
|
NM_010351
|
Gsc
|
goosecoid homeobox
|
|
chr18_+_44264117
|
1.951
|
NM_030061
|
Spink12
|
serine peptidase inhibitor, Kazal type 11
|
|
chr10_+_41250456
|
1.931
|
|
Cd164
|
CD164 antigen
|
|
chr11_+_101848369
|
1.926
|
NM_029287
|
1700006E09Rik
|
RIKEN cDNA 1700006E09 gene
|
|
chrX_+_34784986
|
1.923
|
NM_001085348
NM_029203
|
Rhox2e
Rhox2a
|
reproductive homeobox 2E
reproductive homeobox 2A
|
|
chr17_+_47047407
|
1.922
|
NM_008938
|
Prph2
|
peripherin 2
|
|
chr16_+_37580238
|
1.893
|
NM_013547
|
Hgd
|
homogentisate 1, 2-dioxygenase
|
|
chr17_+_49754460
|
1.877
|
NM_177052
|
Kif6
|
kinesin family member 6
|
|
chr6_+_134874663
|
1.874
|
|
Cdkn1b
|
cyclin-dependent kinase inhibitor 1B
|
|
chr8_+_13037791
|
1.853
|
|
F10
|
coagulation factor X
|
|
chr6_-_98877291
|
1.853
|
|
|
|
|
chrX_-_35057976
|
1.845
|
NM_001114153
|
Rhox2g
|
reproductive homeobox 2G
|
|
chr15_-_66644199
|
1.841
|
|
Sla
|
src-like adaptor
|
|
chr1_-_65149923
|
1.791
|
NM_007774
|
Cryga
|
crystallin, gamma A
|
|
chr11_-_99199267
|
1.776
|
NM_001033397
|
Krt26
|
keratin 26
|
|
chr15_+_39491073
|
1.769
|
|
Rims2
|
regulating synaptic membrane exocytosis 2
|
|
chr18_+_57628139
|
1.765
|
NM_001134697
|
Ctxn3
|
cortexin 3
|
|
chr15_+_80453912
|
1.741
|
NM_010815
|
Grap2
|
GRB2-related adaptor protein 2
|
|
chr4_+_64785795
|
1.720
|
|
Pappa
|
pregnancy-associated plasma protein A
|
|
chr17_-_72102224
|
1.719
|
NM_146082
|
BC027072
|
cDNA sequence BC027072
|
|
chr12_+_53800302
|
1.718
|
|
Akap6
|
A kinase (PRKA) anchor protein 6
|
|
chr18_+_37467108
|
1.715
|
NM_053129
|
Pcdhb4
|
protocadherin beta 4
|
|
chr2_+_163372889
|
1.705
|
NM_008261
|
Hnf4a
|
hepatic nuclear factor 4, alpha
|
|
chr7_+_130267661
|
1.682
|
|
Tnrc6a
|
trinucleotide repeat containing 6a
|
|
chr11_-_33893151
|
1.668
|
NM_001190886
|
Kcnip1
|
Kv channel-interacting protein 1
|
|
chr5_+_140019846
|
1.659
|
NM_013702
|
Uncx
|
UNC homeobox
|
|
chr2_-_165709697
|
1.655
|
|
Zmynd8
|
zinc finger, MYND-type containing 8
|
|
chr15_-_101404334
|
1.649
|
NM_133357
|
Krt75
|
keratin 75
|
|
chr5_+_27588462
|
1.642
|
NM_001136060
|
Dpp6
|
dipeptidylpeptidase 6
|
|
chr10_-_106922986
|
1.639
|
NM_008656
|
Myf5
|
myogenic factor 5
|
|
chr6_+_54251485
|
1.635
|
|
|
|
|
chr15_+_8918001
|
1.634
|
NM_198024
|
Ranbp3l
|
RAN binding protein 3-like
|
|
chr19_-_39723518
|
1.613
|
NM_001024719
|
Cyp2c67
|
cytochrome P450, family 2, subfamily c, polypeptide 67
|
|
chr4_-_126540249
|
1.610
|
|
Zmym4
|
zinc finger, MYM-type 4
|
|
chr7_+_104840013
|
1.601
|
|
Rsf1
|
remodeling and spacing factor 1
|
|
chr2_-_127346987
|
1.599
|
|
Kcnip3
|
Kv channel interacting protein 3, calsenilin
|
|
chr10_+_128143313
|
1.580
|
NM_021882
|
Pmel
|
premelanosome protein
|
|
chr1_-_175157322
|
1.573
|
NM_010184
|
Fcer1a
|
Fc receptor, IgE, high affinity I, alpha polypeptide
|
|
chr1_-_173165051
|
1.572
|
|
|
|
|
chr4_+_133109291
|
1.566
|
NM_011850
|
Nr0b2
|
nuclear receptor subfamily 0, group B, member 2
|
|
chr7_+_86418127
|
1.561
|
NM_018811
|
Abhd2
|
abhydrolase domain containing 2
|
|
chr11_+_94871643
|
1.560
|
|
Samd14
|
sterile alpha motif domain containing 14
|
|
chr9_+_75623899
|
1.549
|
|
Bmp5
|
bone morphogenetic protein 5
|
|
chr1_+_109258889
|
1.529
|
NM_025867
|
Serpinb11
|
serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 11
|
|
chr4_-_129167980
|
1.518
|
NM_080442
|
Tssk3
|
testis-specific serine kinase 3
|
|
chr2_-_27118325
|
1.511
|
|
Vav2
|
vav 2 oncogene
|
|
chr4_+_150302107
|
1.507
|
NM_001077509
|
Tnfrsf9
|
tumor necrosis factor receptor superfamily, member 9
|
|
chr5_+_19413335
|
1.496
|
NM_001170745
NM_015823
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2
|
|
chr19_-_8040489
|
1.493
|
NM_134256
|
AB056442
|
cDNA sequence AB056442
|
|
chr5_-_138628389
|
1.476
|
NM_009315
|
Taf6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor
|
|
chr2_-_167124096
|
1.465
|
|
|
|
|
chr7_-_19895354
|
1.464
|
NM_008036
|
Fosb
|
FBJ osteosarcoma oncogene B
|
|
chr1_+_174486066
|
1.459
|
NM_177723
|
Vsig8
|
V-set and immunoglobulin domain containing 8
|
|
chr8_-_3625510
|
1.458
|
NM_001129803
|
Pcp2
|
Purkinje cell protein 2 (L7)
|
|
chr15_+_58731646
|
1.452
|
|
Rnf139
|
ring finger protein 139
|
|
chr17_-_27304602
|
1.439
|
NM_173027
|
Ip6k3
|
inositol hexaphosphate kinase 3
|
|
chr5_-_52541869
|
1.430
|
|
Dhx15
|
DEAH (Asp-Glu-Ala-His) box polypeptide 15
|
|
chr9_+_118387306
|
1.425
|
NM_001164789
NM_010136
|
Eomes
|
eomesodermin homolog (Xenopus laevis)
|
|
chr2_-_127347102
|
1.423
|
NM_019789
|
Kcnip3
|
Kv channel interacting protein 3, calsenilin
|
|
chr7_+_36234110
|
1.422
|
NM_001199015
NM_001199016
NM_021291
|
Slc7a9
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 9
|
|
chr6_+_13821522
|
1.420
|
|
1110019D14Rik
|
RIKEN cDNA 1110019D14 gene
|
|
chr8_+_4238862
|
1.409
|
|
Map2k7
|
mitogen-activated protein kinase kinase 7
|
|
chr11_-_99471502
|
1.391
|
NM_001099346
|
Gm11937
|
predicted gene 11937
|
|
chr1_-_180259336
|
1.373
|
|
Hnrnpu
|
heterogeneous nuclear ribonucleoprotein U
|
|
chr7_-_52965801
|
1.372
|
|
Sphk2
|
sphingosine kinase 2
|
|
chr16_+_37580303
|
1.371
|
|
Hgd
|
homogentisate 1, 2-dioxygenase
|
|
chr8_+_97197870
|
1.367
|
|
Arl2bp
|
ADP-ribosylation factor-like 2 binding protein
|
|
chr19_+_31943250
|
1.362
|
NM_001081074
|
A1cf
|
APOBEC1 complementation factor
|
|
chr12_+_53800369
|
1.360
|
NM_198111
|
Akap6
|
A kinase (PRKA) anchor protein 6
|
|
chr5_+_37442095
|
1.346
|
NM_178394
|
Jakmip1
|
janus kinase and microtubule interacting protein 1
|
|
chr4_-_136891931
|
1.344
|
|
Cdc42
|
cell division cycle 42 homolog (S. cerevisiae)
|
|
chr10_+_58866370
|
1.343
|
NM_023530
|
Pla2g12b
|
phospholipase A2, group XIIB
|
|
chr5_+_29972960
|
1.334
|
|
Ube3c
|
ubiquitin protein ligase E3C
|
|
chr9_+_32201569
|
1.332
|
NM_001168354
|
Kcnj1
|
potassium inwardly-rectifying channel, subfamily J, member 1
|
|
chr2_-_79296732
|
1.329
|
NM_010894
|
Neurod1
|
neurogenic differentiation 1
|
|
chr1_-_89283173
|
1.322
|
|
|
|
|
chr2_+_9802878
|
1.319
|
|
4930412O13Rik
|
RIKEN cDNA 4930412O13 gene
|
|
chr7_-_94641859
|
1.313
|
NM_011661
|
Tyr
|
tyrosinase
|
|
chr7_+_86418082
|
1.301
|
|
Abhd2
|
abhydrolase domain containing 2
|
|
chr17_+_35670145
|
1.299
|
NM_020576
|
Psors1c2
|
psoriasis susceptibility 1 candidate 2 (human)
|
|
chr3_+_89324085
|
1.298
|
NM_080466
|
Kcnn3
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3
|
|
chr3_-_20142068
|
1.298
|
NM_007753
|
Cpa3
|
carboxypeptidase A3, mast cell
|
|
chr5_+_27588579
|
1.292
|
|
Dpp6
|
dipeptidylpeptidase 6
|
|
chr16_+_20692792
|
1.279
|
|
|
|
|
chr7_+_130267398
|
1.267
|
NM_144925
|
Tnrc6a
|
trinucleotide repeat containing 6a
|
|
chr13_+_8202111
|
1.263
|
NM_052977
|
Adarb2
|
adenosine deaminase, RNA-specific, B2
|
|
chr6_+_57159851
|
1.256
|
NM_053235
|
Vmn1r13
|
vomeronasal 1 receptor 13
|
|
chr9_+_106366204
|
1.255
|
|
|
|
|
chr17_+_74898763
|
1.247
|
|
Yipf4
|
Yip1 domain family, member 4
|
|
chr10_-_116786025
|
1.234
|
|
Cpsf6
|
cleavage and polyadenylation specific factor 6
|
|
chr1_-_129998984
|
1.223
|
NM_027678
|
Zranb3
|
zinc finger, RAN-binding domain containing 3
|