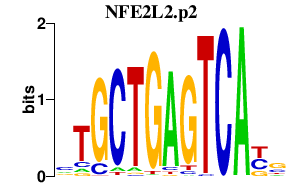
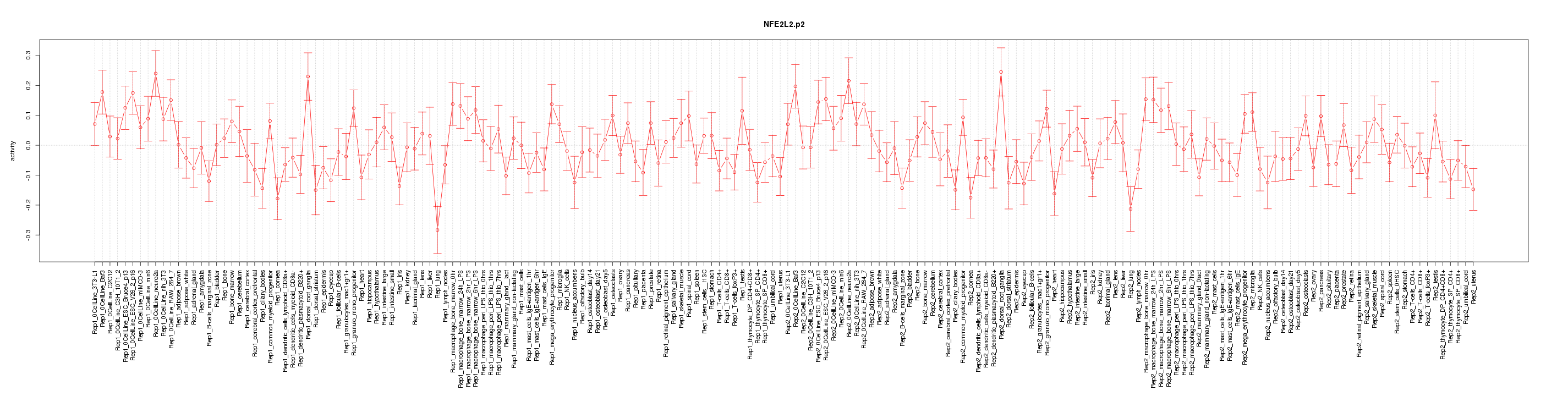
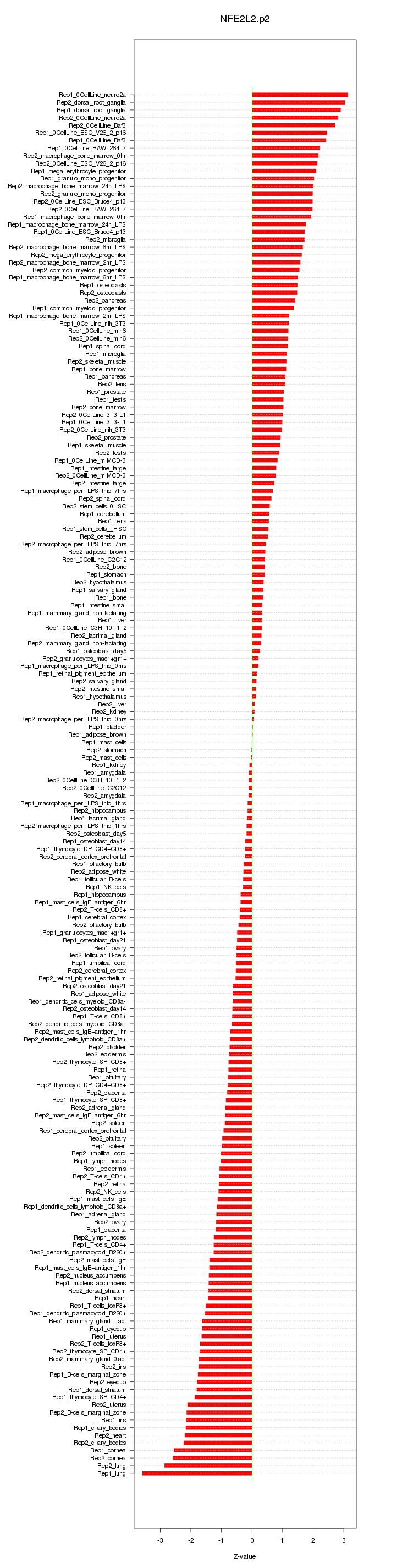
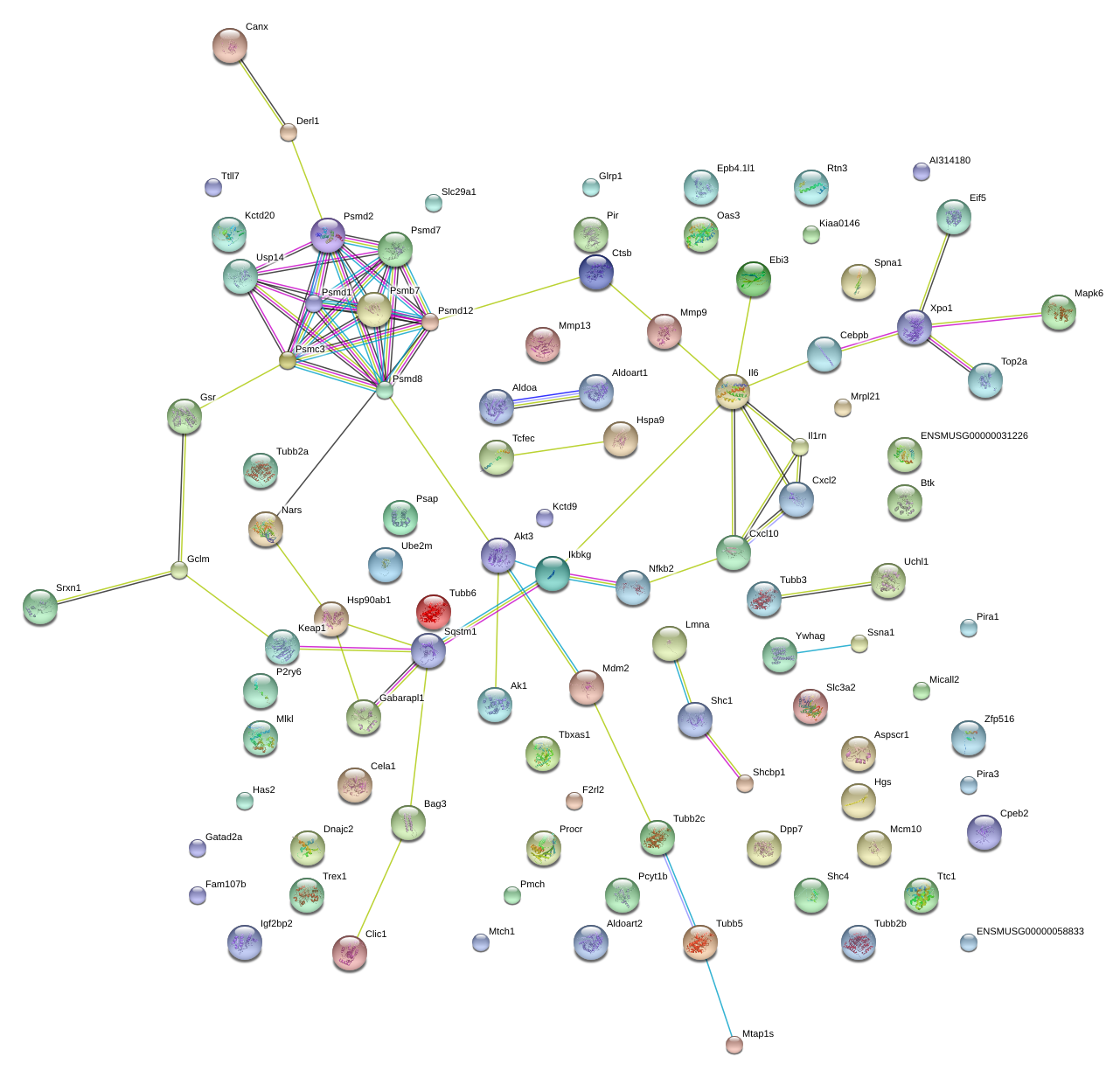
|
chr11_-_98862163
|
17.476
|
|
Top2a
|
topoisomerase (DNA) II alpha
|
|
chr7_-_133942605
|
14.448
|
NM_001177307
|
Aldoa
|
aldolase A, fructose-bisphosphate
|
|
chr2_+_151931424
|
13.990
|
|
Srxn1
|
sulfiredoxin 1 homolog (S. cerevisiae)
|
|
chr7_-_133942767
|
13.675
|
NM_007438
|
Aldoa
|
aldolase A, fructose-bisphosphate
|
|
chr9_+_7272513
|
12.107
|
NM_008607
|
Mmp13
|
matrix metallopeptidase 13
|
|
chr5_-_140212063
|
11.965
|
NM_174850
|
Micall2
|
MICAL-like 2
|
|
chr2_-_25079684
|
11.384
|
|
Tubb2c
|
tubulin, beta 2C
|
|
chr15_-_100517580
|
11.303
|
|
Cela1
|
chymotrypsin-like elastase family, member 1
|
|
chr11_-_43561446
|
11.030
|
NM_133795
|
Ttc1
|
tetratricopeptide repeat domain 1
|
|
chr8_-_110112368
|
10.962
|
NM_010817
|
Psmd7
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 7
|
|
chr5_+_67067497
|
10.957
|
|
Uchl1
|
ubiquitin carboxy-terminal hydrolase L1
|
|
chr5_-_136410488
|
10.685
|
NM_018871
|
Ywhag
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide
|
|
chrX_+_71669985
|
10.628
|
|
Ikbkg
|
inhibitor of kappaB kinase gamma
|
|
chr8_+_34763918
|
10.310
|
|
Gsr
|
glutathione reductase
|
|
chr14_+_63754443
|
10.272
|
|
Ctsb
|
cathepsin B
|
|
chr3_+_121948541
|
10.125
|
|
Gclm
|
glutamate-cysteine ligase, modifier subunit
|
|
chr6_-_16848352
|
10.082
|
NM_031198
|
Tcfec
|
transcription factor EC
|
|
chr2_+_151931465
|
10.001
|
NM_029688
|
Srxn1
|
sulfiredoxin 1 homolog (S. cerevisiae)
|
|
chr3_+_121948693
|
9.805
|
|
Gclm
|
glutamate-cysteine ligase, modifier subunit
|
|
chr11_-_50024275
|
9.545
|
NM_011018
|
Sqstm1
|
sequestosome 1
|
|
chr5_+_30339682
|
9.517
|
NM_031168
|
Il6
|
interleukin 6
|
|
chr3_-_88297220
|
9.356
|
NM_019390
|
Lmna
|
lamin A
|
|
chr9_-_21043503
|
9.082
|
|
Keap1
|
kelch-like ECH-associated protein 1
|
|
chr3_+_121948399
|
8.661
|
NM_008129
|
Gclm
|
glutamate-cysteine ligase, modifier subunit
|
|
chr7_-_29965655
|
8.555
|
NM_026545
|
Psmd8
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 8
|
|
chr3_+_89225539
|
8.551
|
NM_001113331
|
Shc1
|
src homology 2 domain-containing transforming protein C1
|
|
chr17_-_45729199
|
8.458
|
NM_022880
|
Slc29a1
|
solute carrier family 29 (nucleoside transporters), member 1
|
|
chr1_+_87961184
|
8.329
|
NM_027357
|
Psmd1
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 1
|
|
chrX_+_71669971
|
8.277
|
|
Ikbkg
|
inhibitor of kappaB kinase gamma
|
|
chr6_-_16848314
|
8.271
|
|
Tcfec
|
transcription factor EC
|
|
chr9_-_75257067
|
8.113
|
NM_027418
|
Mapk6
|
mitogen-activated protein kinase 6
|
|
chr11_+_107340794
|
8.048
|
NM_025894
|
Psmd12
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 12
|
|
chr2_+_156301543
|
7.969
|
NM_001006664
|
Epb4.1l1
|
erythrocyte protein band 4.1-like 1
|
|
chr2_+_164773719
|
7.940
|
NM_013599
|
Mmp9
|
matrix metallopeptidase 9
|
|
chrX_-_131117602
|
7.929
|
NM_013482
|
Btk
|
Bruton agammaglobulinemia tyrosine kinase
|
|
chr17_+_35187185
|
7.884
|
NM_033444
|
Clic1
|
chloride intracellular channel 1
|
|
chr6_+_38868942
|
7.810
|
NM_011539
|
Tbxas1
|
thromboxane A synthase 1, platelet
|
|
chr16_-_16073490
|
7.636
|
|
2310008H04Rik
|
RIKEN cDNA 2310008H04 gene
|
|
chr18_-_35114005
|
7.530
|
|
Hspa9
|
heat shock protein 9
|
|
chrX_+_160707362
|
7.526
|
NM_027153
|
Pir
|
pirin
|
|
chr16_-_22121346
|
7.367
|
|
Igf2bp2
|
insulin-like growth factor 2 mRNA binding protein 2
|
|
chr11_-_50125157
|
7.197
|
|
Canx
|
calnexin
|
|
chr18_-_35114000
|
7.153
|
|
Hspa9
|
heat shock protein 9
|
|
chr8_+_73435716
|
7.035
|
|
Mtap1s
|
microtubule-associated protein 1S
|
|
chr17_+_35187325
|
7.011
|
|
Clic1
|
chloride intracellular channel 1
|
|
chr5_-_92777875
|
6.710
|
NM_021274
|
Cxcl10
|
chemokine (C-X-C motif) ligand 10
|
|
chr18_-_35114004
|
6.628
|
NM_010481
|
Hspa9
|
heat shock protein 9
|
|
chr9_-_21043246
|
6.594
|
|
Keap1
|
kelch-like ECH-associated protein 1
|
|
chr7_+_135667095
|
6.516
|
NM_013863
|
Bag3
|
BCL2-associated athanogene 3
|
|
chr18_-_35113995
|
6.507
|
|
Hspa9
|
heat shock protein 9
|
|
chr1_+_176102903
|
6.406
|
NM_011465
|
Spna1
|
spectrin alpha 1
|
|
chr8_-_72498125
|
6.286
|
|
Gatad2a
|
GATA zinc finger domain containing 2A
|
|
chr18_-_35113903
|
6.278
|
|
Hspa9
|
heat shock protein 9
|
|
chr7_-_13623158
|
6.244
|
NM_145578
|
Ube2m
|
ubiquitin-conjugating enzyme E2M (UBC12 homolog, yeast)
|
|
chr11_+_120534337
|
6.130
|
|
Aspscr1
|
alveolar soft part sarcoma chromosome region, candidate 1 (human)
|
|
chr19_-_7557553
|
6.116
|
|
Rtn3
|
reticulon 3
|
|
chr17_-_45710161
|
6.084
|
NM_008302
|
Hsp90ab1
|
heat shock protein 90 alpha (cytosolic), class B member 1
|
|
chr5_-_21290925
|
6.010
|
|
Dnajc2
|
DnaJ (Hsp40) homolog, subfamily C, member 2
|
|
chr2_-_125549883
|
5.933
|
NM_199022
|
Shc4
|
SHC (Src homology 2 domain containing) family, member 4
|
|
chr1_-_179178874
|
5.931
|
NM_011785
|
Akt3
|
thymoma viral proto-oncogene 3
|
|
chr2_+_3630817
|
5.915
|
|
Fam107b
|
family with sequence similarity 107, member B
|
|
chr10_-_117147068
|
5.866
|
|
Mdm2
|
transformed mouse 3T3 cell double minute 2
|
|
chr18_-_10029925
|
5.823
|
NM_001038589
NM_021522
|
Usp14
|
ubiquitin specific peptidase 14
|
|
chr8_-_73030573
|
5.750
|
NM_025577
|
2810428I15Rik
|
RIKEN cDNA 2810428I15 gene
|
|
chr11_+_23156482
|
5.704
|
|
Xpo1
|
exportin 1, CRM1 homolog (yeast)
|
|
chr16_+_20651724
|
5.612
|
NM_134101
|
Psmd2
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 2
|
|
chr2_+_155576833
|
5.583
|
|
Procr
|
protein C receptor, endothelial
|
|
chr11_+_120494714
|
5.551
|
|
|
|
|
chr9_-_21043375
|
5.520
|
|
Keap1
|
kelch-like ECH-associated protein 1
|
|
chr2_+_156301651
|
5.419
|
|
Epb4.1l1
|
erythrocyte protein band 4.1-like 1
|
|
chr17_-_45710213
|
5.418
|
|
Hsp90ab1
|
heat shock protein 90 alpha (cytosolic), class B member 1
|
|
chr2_-_4912152
|
5.382
|
|
Mcm10
|
minichromosome maintenance deficient 10 (S. cerevisiae)
|
|
chr7_-_108112785
|
5.352
|
NM_183168
|
P2ry6
|
pyrimidinergic receptor P2Y, G-protein coupled, 6
|
|
chrX_+_71669933
|
5.314
|
NM_001136067
NM_001161421
|
Ikbkg
|
inhibitor of kappaB kinase gamma
|
|
chrX_+_102275071
|
5.298
|
NM_026312
|
2610029G23Rik
|
RIKEN cDNA 2610029G23 gene
|
|
chr9_-_21043659
|
5.278
|
NM_001110305
NM_001110306
NM_001110307
NM_016679
|
Keap1
|
kelch-like ECH-associated protein 1
|
|
chr2_+_24192373
|
5.257
|
NM_031167
|
Il1rn
|
interleukin 1 receptor antagonist
|
|
chr2_+_90894747
|
5.244
|
|
Psmc3
|
proteasome (prosome, macropain) 26S subunit, ATPase 3
|
|
chr2_+_167514489
|
5.207
|
|
Cebpb
|
CCAAT/enhancer binding protein (C/EBP), beta
|
|
chr5_+_91332895
|
5.019
|
NM_009140
|
Cxcl2
|
chemokine (C-X-C motif) ligand 2
|
|
chr5_-_121227617
|
4.983
|
NM_145226
|
Oas3
|
2'-5' oligoadenylate synthetase 3
|
|
chr10_+_59740373
|
4.951
|
NM_001146120
NM_001146121
NM_001146122
NM_001146123
NM_011179
|
Psap
|
prosaposin
|
|
chr17_+_56092075
|
4.925
|
|
Ebi3
|
Epstein-Barr virus induced gene 3
|
|
chr2_-_38499436
|
4.888
|
|
Psmb7
|
proteasome (prosome, macropain) subunit, beta type 7
|
|
chr17_+_29089357
|
4.852
|
|
Kctd20
|
potassium channel tetramerisation domain containing 20
|
|
chr19_-_8787823
|
4.765
|
|
Slc3a2
|
solute carrier family 3 (activators of dibasic and neutral amino acid transport), member 2
|
|
chr17_+_29089623
|
4.694
|
|
Kctd20
|
potassium channel tetramerisation domain containing 20
|
|
chr8_-_4779533
|
4.666
|
NM_011369
|
Shcbp1
|
Shc SH2-domain binding protein 1
|
|
chr11_+_120328948
|
4.662
|
NM_001159328
NM_008244
|
Hgs
|
HGF-regulated tyrosine kinase substrate
|
|
chr14_+_68334187
|
4.657
|
|
Kctd9
|
potassium channel tetramerisation domain containing 9
|
|
chr8_-_113862056
|
4.625
|
|
Mlkl
|
mixed lineage kinase domain-like
|
|
chr1_-_90406613
|
4.624
|
NM_008132
|
Glrp1
|
glutamine repeat protein 1
|
|
chr6_+_129483154
|
4.583
|
|
Gabarapl1
|
gamma-aminobutyric acid (GABA) A receptor-associated protein-like 1
|
|
chr2_-_25080099
|
4.581
|
|
Tubb2c
|
tubulin, beta 2C
|
|
chr8_-_73030643
|
4.564
|
|
2810428I15Rik
|
RIKEN cDNA 2810428I15 gene
|
|
chr15_-_56526052
|
4.514
|
NM_008216
|
Has2
|
hyaluronan synthase 2
|
|
chr5_-_21290971
|
4.498
|
NM_009584
|
Dnajc2
|
DnaJ (Hsp40) homolog, subfamily C, member 2
|
|
chr18_+_83080270
|
4.452
|
NM_183033
|
Zfp516
|
zinc finger protein 516
|
|
chr2_-_25210076
|
4.438
|
|
Dpp7
|
dipeptidylpeptidase 7
|
|
chr7_+_135667279
|
4.405
|
|
Bag3
|
BCL2-associated athanogene 3
|
|
chr2_-_38499375
|
4.365
|
NM_011187
|
Psmb7
|
proteasome (prosome, macropain) subunit, beta type 7
|
|
chr15_-_57702643
|
4.364
|
|
Derl1
|
Der1-like domain family, member 1
|
|
chr4_-_58925575
|
4.352
|
NM_172381
|
AI314180
|
expressed sequence AI314180
|
|
chr12_+_112776828
|
4.336
|
|
Eif5
|
eukaryotic translation initiation factor 5
|
|
chr3_+_146515330
|
4.269
|
NM_027594
|
Ttll7
|
tubulin tyrosine ligase-like family, member 7
|
|
chr18_-_64676144
|
4.255
|
|
Nars
|
asparaginyl-tRNA synthetase
|
|
chrX_+_90920450
|
4.247
|
NM_211138
|
Pcyt1b
|
phosphate cytidylyltransferase 1, choline, beta isoform
|
|
chr6_+_129483238
|
4.142
|
|
Gabarapl1
|
gamma-aminobutyric acid (GABA) A receptor-associated protein-like 1
|
|
chr2_+_32483909
|
4.111
|
NM_001198792
|
Ak1
|
adenylate kinase 1
|
|
chr2_-_25127860
|
3.987
|
NM_023464
|
Ssna1
|
Sjogren's syndrome nuclear autoantigen 1
|
|
chr10_+_87553816
|
3.964
|
NM_029971
|
Pmch
|
pro-melanin-concentrating hormone
|
|
chr9_-_108962219
|
3.962
|
NM_011637
|
Trex1
|
three prime repair exonuclease 1
|
|
chr2_+_90894161
|
3.905
|
NM_008948
|
Psmc3
|
proteasome (prosome, macropain) 26S subunit, ATPase 3
|
|
chr10_+_59740430
|
3.805
|
|
Psap
|
prosaposin
|
|
chr8_-_110112260
|
3.775
|
|
Psmd7
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 7
|
|
chr11_+_72263260
|
3.742
|
|
|
|
|
chr13_+_96466807
|
3.695
|
NM_010170
|
F2rl2
|
coagulation factor II (thrombin) receptor-like 2
|
|
chr5_+_43624701
|
3.681
|
NM_001177379
NM_175937
|
Cpeb2
|
cytoplasmic polyadenylation element binding protein 2
|
|
chr19_+_3283038
|
3.608
|
NM_172252
|
Mrpl21
|
mitochondrial ribosomal protein L21
|
|
chr19_+_46380418
|
3.603
|
NM_001177370
|
Nfkb2
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2, p49/p100
|
|
chr18_-_63851841
|
3.568
|
NM_016792
|
Txnl1
|
thioredoxin-like 1
|
|
chr9_-_108961721
|
3.530
|
NM_001012236
|
Trex1
|
three prime repair exonuclease 1
|
|
chr6_+_34304098
|
3.504
|
NM_008012
|
Akr1b8
|
aldo-keto reductase family 1, member B8
|
|
chr11_-_109334909
|
3.491
|
NM_001029842
|
Slc16a6
|
solute carrier family 16 (monocarboxylic acid transporters), member 6
|
|
chr2_+_11627385
|
3.490
|
NM_008358
|
Il15ra
|
interleukin 15 receptor, alpha chain
|
|
chr6_+_48986501
|
3.485
|
NM_053110
|
Gpnmb
|
glycoprotein (transmembrane) nmb
|
|
chr10_+_110910277
|
3.452
|
|
Nap1l1
|
nucleosome assembly protein 1-like 1
|
|
chr6_-_83267540
|
3.434
|
NM_008638
|
Mthfd2
|
methylenetetrahydrofolate dehydrogenase (NAD+ dependent), methenyltetrahydrofolate cyclohydrolase
|
|
chr7_+_52104591
|
3.421
|
NM_026270
|
Akt1s1
|
AKT1 substrate 1 (proline-rich)
|
|
chr11_-_87800086
|
3.416
|
NM_001168472
|
Dynll2
|
dynein light chain LC8-type 2
|
|
chr13_-_58316923
|
3.367
|
|
Ubqln1
|
ubiquilin 1
|
|
chr19_-_7557690
|
3.362
|
NM_053076
NM_001003933
NM_001003934
|
Rtn3
|
reticulon 3
|
|
chr18_-_64676090
|
3.362
|
|
Nars
|
asparaginyl-tRNA synthetase
|
|
chr16_+_32430986
|
3.362
|
NM_009981
NM_001163159
|
Pcyt1a
|
phosphate cytidylyltransferase 1, choline, alpha isoform
|
|
chr3_+_146515369
|
3.360
|
|
Ttll7
|
tubulin tyrosine ligase-like family, member 7
|
|
chrX_-_99352341
|
3.301
|
NM_146235
|
Ercc6l
|
excision repair cross-complementing rodent repair deficiency complementation group 6 - like
|
|
chr6_+_145756902
|
3.287
|
NM_027760
|
Rassf8
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8
|
|
chr18_-_64676160
|
3.263
|
NM_001142950
NM_027350
|
Nars
|
asparaginyl-tRNA synthetase
|
|
chr11_-_100565382
|
3.246
|
NM_030150
|
Dhx58
|
DEXH (Asp-Glu-X-His) box polypeptide 58
|
|
chr17_+_28829512
|
3.226
|
NM_001168513
|
Mapk14
|
mitogen-activated protein kinase 14
|
|
chr11_-_120979990
|
3.187
|
NM_027622
|
Tex19.2
|
testis expressed gene 19.2
|
|
chr1_-_79436564
|
3.172
|
NM_009129
|
Scg2
|
secretogranin II
|
|
chr12_-_113205172
|
3.127
|
NM_027360
|
2010107E04Rik
|
RIKEN cDNA 2010107E04 gene
|
|
chr3_+_108060061
|
3.112
|
|
Psma5
|
proteasome (prosome, macropain) subunit, alpha type 5
|
|
chr12_-_113205152
|
3.104
|
|
2010107E04Rik
|
RIKEN cDNA 2010107E04 gene
|
|
chr7_+_5996762
|
3.102
|
NM_031389
|
Nlrp4c
|
NLR family, pyrin domain containing 4C
|
|
chr7_-_35015309
|
3.031
|
NM_008155
|
Gpi1
|
glucose phosphate isomerase 1
|
|
chr11_-_120298946
|
3.005
|
NM_001195023
NM_199469
|
Nploc4
|
nuclear protein localization 4 homolog (S. cerevisiae)
|
|
chr13_+_75977323
|
2.981
|
NM_053108
|
Glrx
|
glutaredoxin
|
|
chr2_+_155576943
|
2.975
|
NM_011171
|
Procr
|
protein C receptor, endothelial
|
|
chr11_+_121007456
|
2.961
|
NM_028602
|
Tex19.1
|
testis expressed gene 19.1
|
|
chr4_-_57969277
|
2.960
|
NM_011660
|
Txn1
|
thioredoxin 1
|
|
chr7_+_148654681
|
2.959
|
NM_001111050
|
Cd151
|
CD151 antigen
|
|
chr7_-_30980042
|
2.950
|
NM_009795
|
Capns1
|
calpain, small subunit 1
|
|
chr8_-_129469539
|
2.945
|
|
Tomm20
|
translocase of outer mitochondrial membrane 20 homolog (yeast)
|
|
chr2_+_90894434
|
2.926
|
|
Psmc3
|
proteasome (prosome, macropain) 26S subunit, ATPase 3
|
|
chr14_+_68334154
|
2.925
|
NM_001111028
NM_134073
|
Kctd9
|
potassium channel tetramerisation domain containing 9
|
|
chrX_+_156278399
|
2.899
|
NM_001135728
|
Sh3kbp1
|
SH3-domain kinase binding protein 1
|
|
chrX_+_12858142
|
2.890
|
NM_010028
|
Ddx3x
|
DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 3, X-linked
|
|
chr5_-_109104366
|
2.882
|
|
Slc26a1
|
solute carrier family 26 (sulfate transporter), member 1
|
|
chr8_+_125372273
|
2.860
|
NM_007662
|
Cdh15
|
cadherin 15
|
|
chr5_-_109104394
|
2.860
|
|
Slc26a1
|
solute carrier family 26 (sulfate transporter), member 1
|
|
chr8_+_96696512
|
2.821
|
NM_008630
|
Mt2
|
metallothionein 2
|
|
chr6_+_48986625
|
2.808
|
|
Gpnmb
|
glycoprotein (transmembrane) nmb
|
|
chr14_-_21212834
|
2.805
|
|
Mrps16
|
mitochondrial ribosomal protein S16
|
|
chr7_+_31338319
|
2.798
|
NM_001012401
|
Hspb6
|
heat shock protein, alpha-crystallin-related, B6
|
|
chr13_-_64471609
|
2.778
|
NM_009984
|
Ctsl
|
cathepsin L
|
|
chr7_-_148202779
|
2.767
|
NM_001033632
|
Ifitm6
|
interferon induced transmembrane protein 6
|
|
chr1_+_109486582
|
2.743
|
NM_001159748
NM_011459
|
Serpinb8
|
serine (or cysteine) peptidase inhibitor, clade B, member 8
|
|
chr8_+_112302530
|
2.732
|
NM_009677
|
Ap1g1
|
adaptor protein complex AP-1, gamma 1 subunit
|
|
chr11_-_69575192
|
2.719
|
NM_019871
|
Amac1
|
acyl-malonyl condensing enzyme 1
|
|
chr12_-_77896534
|
2.719
|
NM_030677
|
Gpx2
|
glutathione peroxidase 2
|
|
chr7_-_35015297
|
2.693
|
|
Gpi1
|
glucose phosphate isomerase 1
|
|
chr7_+_117916117
|
2.677
|
NM_009667
|
Ampd3
|
adenosine monophosphate deaminase 3
|
|
chr10_+_79727708
|
2.666
|
NM_001122604
NM_001122605
NM_133188
|
Dazap1
|
DAZ associated protein 1
|
|
chr5_-_100929644
|
2.660
|
NM_172714
|
Lin54
|
lin-54 homolog (C. elegans)
|
|
chr3_+_94926272
|
2.617
|
|
|
|
|
chr11_-_114795651
|
2.603
|
NM_199221
|
Cd300lb
|
CD300 antigen like family member B
|
|
chrX_-_140261831
|
2.575
|
|
Capn6
|
calpain 6
|
|
chr11_-_74991244
|
2.574
|
|
Ovca2
|
candidate tumor suppressor in ovarian cancer 2
|
|
chr5_-_109104347
|
2.530
|
|
Slc26a1
|
solute carrier family 26 (sulfate transporter), member 1
|
|
chr2_+_121854287
|
2.525
|
NM_144545
|
Eif3j
|
eukaryotic translation initiation factor 3, subunit J
|
|
chr8_+_34763709
|
2.510
|
NM_010344
|
Gsr
|
glutathione reductase
|
|
chr2_-_163245205
|
2.504
|
NM_025699
|
3230401D17Rik
|
RIKEN cDNA 3230401D17 gene
|
|
chr12_-_77896363
|
2.495
|
|
Gpx2
|
glutathione peroxidase 2
|
|
chr3_-_89151364
|
2.482
|
|
Adam15
|
a disintegrin and metallopeptidase domain 15 (metargidin)
|
|
chr18_+_12802276
|
2.465
|
|
Ttc39c
|
tetratricopeptide repeat domain 39C
|
|
chr12_+_112997194
|
2.444
|
|
Klc1
|
kinesin light chain 1
|
|
chr3_+_28704426
|
2.440
|
NM_026517
|
Rpl22l1
|
ribosomal protein L22 like 1
|
|
chr3_+_28704357
|
2.420
|
|
Rpl22l1
|
ribosomal protein L22 like 1
|
|
chrX_-_140261954
|
2.405
|
NM_007603
|
Capn6
|
calpain 6
|
|
chr16_-_22439642
|
2.402
|
NM_023794
|
Etv5
|
ets variant gene 5
|
|
chr1_+_137914574
|
2.397
|
NM_025439
|
Tmem9
|
transmembrane protein 9
|
|
chr2_-_162915336
|
2.395
|
NM_026630
|
Gtsf1l
|
gametocyte specific factor 1-like
|
|
chr11_-_115884885
|
2.387
|
|
|
|
|
chr13_-_19711409
|
2.383
|
NM_134065
|
Epdr1
|
ependymin related protein 1 (zebrafish)
|
|
chr1_-_31006599
|
2.373
|
NM_011200
|
Ptp4a1
|
protein tyrosine phosphatase 4a1
|
|
chr7_+_86058650
|
2.362
|
NM_020583
|
Isg20
|
interferon-stimulated protein
|
|
chr7_+_134388553
|
2.356
|
|
Zfp771
|
zinc finger protein 771
|
|
chr3_-_107877497
|
2.353
|
|
Ampd2
|
adenosine monophosphate deaminase 2
|