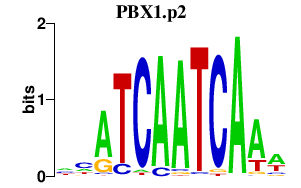
|
chr9_+_65738168
|
29.416
|
|
2810417H13Rik
|
RIKEN cDNA 2810417H13 gene
|
|
chr9_+_65738150
|
27.927
|
|
2810417H13Rik
|
RIKEN cDNA 2810417H13 gene
|
|
chr5_+_111268995
|
26.002
|
|
Chek2
|
CHK2 checkpoint homolog (S. pombe)
|
|
chr13_-_21925343
|
25.189
|
NM_178185
NM_001177544
|
Hist1h2ah
Hist1h2ap
Hist1h2ai
Hist1h2ao
|
histone cluster 1, H2ah
histone cluster 1, H2ap
histone cluster 1, H2ai
histone cluster 1, H2ao
|
|
chr13_+_23666249
|
24.790
|
NM_178188
|
Hist1h2ad
Hist1h2ai
|
histone cluster 1, H2ad
histone cluster 1, H2ai
|
|
chr9_+_65738129
|
24.010
|
NM_026515
|
2810417H13Rik
|
RIKEN cDNA 2810417H13 gene
|
|
chr8_-_4779533
|
23.719
|
NM_011369
|
Shcbp1
|
Shc SH2-domain binding protein 1
|
|
chr2_-_172195998
|
21.695
|
NM_011497
|
Aurka
|
aurora kinase A
|
|
chr13_-_23775827
|
21.668
|
NM_178189
|
Hist1h2ac
|
histone cluster 1, H2ac
|
|
chr5_+_111269020
|
21.460
|
NM_016681
|
Chek2
|
CHK2 checkpoint homolog (S. pombe)
|
|
chr2_+_164595447
|
20.463
|
|
Ube2c
|
ubiquitin-conjugating enzyme E2C
|
|
chr2_+_164595415
|
18.181
|
NM_026785
|
Ube2c
|
ubiquitin-conjugating enzyme E2C
|
|
chr16_-_15637390
|
14.150
|
NM_008565
|
Mcm4
|
minichromosome maintenance deficient 4 homolog (S. cerevisiae)
|
|
chr11_-_40568524
|
13.644
|
|
Ccng1
|
cyclin G1
|
|
chr5_-_111268743
|
13.223
|
NM_153571
|
Hscb
|
HscB iron-sulfur cluster co-chaperone homolog (E. coli)
|
|
chr11_-_40568710
|
12.710
|
NM_009831
|
Ccng1
|
cyclin G1
|
|
chr8_+_59990639
|
12.140
|
NM_008252
|
Hmgb2
|
high mobility group box 2
|
|
chrX_-_71470766
|
11.998
|
|
Flna
|
filamin, alpha
|
|
chr1_-_45559866
|
11.715
|
|
Col5a2
|
collagen, type V, alpha 2
|
|
chr1_-_45559904
|
11.439
|
|
Col5a2
|
collagen, type V, alpha 2
|
|
chr6_-_3349415
|
11.437
|
|
Samd9l
|
sterile alpha motif domain containing 9-like
|
|
chr14_-_68333897
|
11.132
|
NM_175384
|
Cdca2
|
cell division cycle associated 2
|
|
chr10_-_111434265
|
10.730
|
NM_028608
|
Glipr1
|
GLI pathogenesis-related 1 (glioma)
|
|
chr4_+_132324212
|
10.690
|
NM_011284
|
Rpa2
|
replication protein A2
|
|
chr4_+_140257340
|
10.601
|
NM_173867
|
Rcc2
|
regulator of chromosome condensation 2
|
|
chr2_+_164595389
|
10.570
|
|
Ube2c
|
ubiquitin-conjugating enzyme E2C
|
|
chr14_+_68334154
|
10.526
|
NM_001111028
NM_134073
|
Kctd9
|
potassium channel tetramerisation domain containing 9
|
|
chr6_-_8209135
|
10.282
|
NM_026632
|
Rpa3
|
replication protein A3
|
|
chr6_-_3349470
|
10.178
|
NM_010156
|
Samd9l
|
sterile alpha motif domain containing 9-like
|
|
chr5_-_138613028
|
10.007
|
NM_008568
|
Mcm7
|
minichromosome maintenance deficient 7 (S. cerevisiae)
|
|
chr14_+_47993487
|
9.988
|
NM_001145953
NM_010705
|
Lgals3
|
lectin, galactose binding, soluble 3
|
|
chr16_+_94791713
|
9.053
|
NM_007890
|
Dyrk1a
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1a
|
|
chr16_+_30065419
|
9.045
|
NM_008235
|
Hes1
|
hairy and enhancer of split 1 (Drosophila)
|
|
chr6_-_8209105
|
9.034
|
|
Rpa3
|
replication protein A3
|
|
chr19_-_9973901
|
9.023
|
NM_016692
|
Incenp
|
inner centromere protein
|
|
chr1_-_45559700
|
8.899
|
|
Col5a2
|
collagen, type V, alpha 2
|
|
chr12_+_77471506
|
8.812
|
|
Zbtb1
|
zinc finger and BTB domain containing 1
|
|
chr17_-_25864310
|
8.350
|
NM_145409
|
Chtf18
|
CTF18, chromosome transmission fidelity factor 18 homolog (S. cerevisiae)
|
|
chr15_-_99200868
|
7.907
|
NM_011711
|
Fmnl3
|
formin-like 3
|
|
chr9_+_109778051
|
7.741
|
NM_007658
|
Cdc25a
|
cell division cycle 25 homolog A (S. pombe)
|
|
chr4_-_147016606
|
7.557
|
NM_145078
|
2610305D13Rik
|
RIKEN cDNA 2610305D13 gene
|
|
chr15_+_102236709
|
7.262
|
NM_013672
|
Sp1
|
trans-acting transcription factor 1
|
|
chr3_+_102799653
|
6.891
|
|
Sike1
|
suppressor of IKBKE 1
|
|
chr5_-_115608153
|
6.834
|
NM_175403
|
Mlec
|
malectin
|
|
chr10_-_111434231
|
6.814
|
|
Glipr1
|
GLI pathogenesis-related 1 (glioma)
|
|
chr4_+_146124224
|
6.718
|
NM_001177546
|
Zfp600
|
zinc finger protein 600
|
|
chr13_-_23790310
|
6.614
|
NM_178208
|
Hist1h4c
|
histone cluster 1, H4c
|
|
chr13_-_120197722
|
6.525
|
NM_008710
|
Nnt
|
nicotinamide nucleotide transhydrogenase
|
|
chr4_-_106812064
|
6.511
|
|
2210012G02Rik
|
RIKEN cDNA 2210012G02 gene
|
|
chr13_-_23854226
|
6.437
|
NM_013550
|
Hist1h3a
|
histone cluster 1, H3a
|
|
chr3_-_27052794
|
6.356
|
NM_001177626
|
Ect2
|
ect2 oncogene
|
|
chr14_+_68334187
|
6.336
|
|
Kctd9
|
potassium channel tetramerisation domain containing 9
|
|
chr15_-_42508294
|
6.251
|
NM_009640
|
Angpt1
|
angiopoietin 1
|
|
chr4_+_62050088
|
6.247
|
|
Slc31a1
|
solute carrier family 31, member 1
|
|
chr11_-_97023157
|
6.145
|
|
Kpnb1
|
karyopherin (importin) beta 1
|
|
chr4_+_146927385
|
6.141
|
NM_001014397
|
Gm13154
|
predicted gene 13154
|
|
chr9_-_13631343
|
6.126
|
|
Cep57
|
centrosomal protein 57
|
|
chr2_+_119177025
|
6.092
|
|
Chac1
|
ChaC, cation transport regulator-like 1 (E. coli)
|
|
chr17_+_24665193
|
6.038
|
|
2610019E17Rik
|
RIKEN cDNA 2610019E17 gene
|
|
chr6_-_52114644
|
5.992
|
NM_010451
|
Hoxa2
|
homeobox A2
|
|
chr11_-_79894401
|
5.955
|
NM_018776
|
Crlf3
|
cytokine receptor-like factor 3
|
|
chr6_-_3295980
|
5.904
|
|
9530028C05
|
hypothetical protein 9530028C05
|
|
chr6_-_87793459
|
5.862
|
|
Cnbp
|
cellular nucleic acid binding protein
|
|
chr7_-_100022714
|
5.856
|
NM_001080995
|
4632434I11Rik
|
RIKEN cDNA 4632434I11 gene
|
|
chr5_+_31216131
|
5.834
|
NM_133918
|
Emilin1
|
elastin microfibril interfacer 1
|
|
chr6_+_116123984
|
5.801
|
|
Gm8203
|
predicted pseudogene 8203
|
|
chr5_-_123271207
|
5.769
|
|
Anapc5
|
anaphase-promoting complex subunit 5
|
|
chr6_+_83276009
|
5.725
|
NM_145571
|
Mobkl1b
|
MOB1, Mps One Binder kinase activator-like 1B (yeast)
|
|
chr2_+_119176963
|
5.596
|
NM_026929
|
Chac1
|
ChaC, cation transport regulator-like 1 (E. coli)
|
|
chr9_-_13631432
|
5.541
|
|
Cep57
|
centrosomal protein 57
|
|
chr11_+_87895626
|
5.487
|
|
Vezf1
|
vascular endothelial zinc finger 1
|
|
chr1_+_138521477
|
5.416
|
NM_001160251
NM_177643
|
Zfp281
|
zinc finger protein 281
|
|
chr10_-_20880559
|
5.393
|
NM_001198914
NM_010848
|
Myb
|
myeloblastosis oncogene
|
|
chr18_+_67959756
|
5.385
|
NM_027556
|
Cep192
|
centrosomal protein 192
|
|
chr17_+_34375847
|
5.374
|
NM_010389
|
H2-Ob
|
histocompatibility 2, O region beta locus
|
|
chr4_+_145850602
|
5.343
|
NM_001177767
NM_009051
|
Gm13138
Rex2
|
predicted gene 13138
reduced expression 2
|
|
chr4_+_146395958
|
5.343
|
NM_001177767
NM_009051
|
Gm13138
Rex2
|
predicted gene 13138
reduced expression 2
|
|
chr11_-_99464621
|
5.232
|
NM_001127354
|
Gm11938
|
predicted gene 11938
|
|
chr14_-_48037815
|
5.211
|
|
Dlgap5
|
discs, large (Drosophila) homolog-associated protein 5
|
|
chr13_+_81922310
|
5.192
|
|
Cetn3
|
centrin 3
|
|
chr9_-_100446444
|
5.190
|
NM_010878
|
Nck1
|
non-catalytic region of tyrosine kinase adaptor protein 1
|
|
chr4_+_145260599
|
5.168
|
NM_001103158
|
Gm13242
|
predicted gene 13242
|
|
chr17_+_35404109
|
5.136
|
|
|
|
|
chr11_+_96132643
|
4.950
|
NM_008270
|
Hoxb9
|
homeobox B9
|
|
chr2_-_69044179
|
4.948
|
NM_025565
|
Spc25
|
SPC25, NDC80 kinetochore complex component, homolog (S. cerevisiae)
|
|
chr1_-_45560001
|
4.938
|
NM_007737
|
Col5a2
|
collagen, type V, alpha 2
|
|
chr8_+_108424267
|
4.935
|
NM_173432
|
Pskh1
|
protein serine kinase H1
|
|
chr11_-_106860803
|
4.834
|
NM_010655
|
Kpna2
|
karyopherin (importin) alpha 2
|
|
chr5_+_31216069
|
4.825
|
|
Emilin1
|
elastin microfibril interfacer 1
|
|
chr17_+_35403998
|
4.784
|
|
|
|
|
chr1_+_40497472
|
4.757
|
NM_001025602
|
Il1rl1
|
interleukin 1 receptor-like 1
|
|
chr10_-_62666659
|
4.735
|
NM_182992
|
Mypn
|
myopalladin
|
|
chr6_-_39370221
|
4.653
|
|
Mkrn1
|
makorin, ring finger protein, 1
|
|
chr11_-_75114563
|
4.633
|
|
Rpa1
|
replication protein A1
|
|
chr13_+_23627286
|
4.618
|
NM_145073
|
Hist1h3g
|
histone cluster 1, H3g
|
|
chr5_-_123271283
|
4.612
|
NM_001042491
NM_021505
|
Anapc5
|
anaphase-promoting complex subunit 5
|
|
chr8_+_109459623
|
4.551
|
NM_009229
|
Sntb2
|
syntrophin, basic 2
|
|
chr1_+_133424461
|
4.509
|
|
Fam72a
|
family with sequence similarity 72, member A
|
|
chr3_-_27052733
|
4.482
|
NM_001177625
NM_007900
|
Ect2
|
ect2 oncogene
|
|
chr6_-_115803305
|
4.454
|
NM_010774
|
Mbd4
|
methyl-CpG binding domain protein 4
|
|
chr6_-_87793898
|
4.451
|
|
Cnbp
|
cellular nucleic acid binding protein
|
|
chr5_-_115608087
|
4.422
|
|
Mlec
|
malectin
|
|
chr11_+_55283145
|
4.412
|
|
G3bp1
|
Ras-GTPase-activating protein SH3-domain binding protein 1
|
|
chr11_+_55283201
|
4.392
|
NM_013716
|
G3bp1
|
Ras-GTPase-activating protein SH3-domain binding protein 1
|
|
chr13_+_23646900
|
4.384
|
NM_145713
|
Hist1h1d
|
histone cluster 1, H1d
|
|
chr1_-_182743632
|
4.375
|
NM_008210
|
H3f3a
|
H3 histone, family 3A
|
|
chr9_+_109778461
|
4.289
|
|
Cdc25a
|
cell division cycle 25 homolog A (S. pombe)
|
|
chr13_-_23663006
|
4.274
|
NM_178187
|
Hist1h2ae
|
histone cluster 1, H2ae
|
|
chr2_-_3430013
|
4.244
|
NM_015765
|
Hspa14
|
heat shock protein 14
|
|
chr5_+_33715497
|
4.225
|
|
|
|
|
chr2_-_25080099
|
4.205
|
|
Tubb2c
|
tubulin, beta 2C
|
|
chr15_+_102126735
|
4.176
|
|
Espl1
|
extra spindle poles-like 1 (S. cerevisiae)
|
|
chr9_+_78463352
|
4.149
|
NM_153098
|
Cd109
|
CD109 antigen
|
|
chr9_-_87625991
|
4.111
|
NM_023814
|
Tbx18
|
T-box18
|
|
chr2_+_119853216
|
4.081
|
NM_001114637
|
Pla2g4b
Jmjd7
|
phospholipase A2, group IVB (cytosolic)
jumonji domain containing 7
|
|
chrX_+_130842622
|
4.014
|
|
Cenpi
|
centromere protein I
|
|
chr3_+_32607582
|
3.990
|
|
Actl6a
|
actin-like 6A
|
|
chr1_+_95405907
|
3.962
|
|
|
|
|
chr6_-_41654290
|
3.960
|
NM_032540
|
Kel
|
Kell blood group
|
|
chr13_-_21872421
|
3.959
|
NM_020034
|
Hist1h1b
|
histone cluster 1, H1b
|
|
chr8_-_86628606
|
3.957
|
NM_172502
|
Dcaf15
|
DDB1 and CUL4 associated factor 15
|
|
chr8_+_123640069
|
3.852
|
NM_013519
|
Foxc2
|
forkhead box C2
|
|
chr15_-_99200748
|
3.702
|
|
Fmnl3
|
formin-like 3
|
|
chr12_-_32635061
|
3.696
|
|
Hbp1
|
high mobility group box transcription factor 1
|
|
chr14_-_68333666
|
3.674
|
NM_001110162
|
Cdca2
|
cell division cycle associated 2
|
|
chr6_-_125141525
|
3.651
|
NM_146171
|
Ncapd2
|
non-SMC condensin I complex, subunit D2
|
|
chr13_-_23654226
|
3.622
|
NM_178205
|
Hist1h3e
|
histone cluster 1, H3e
|
|
chr13_+_81922236
|
3.619
|
NM_007684
|
Cetn3
|
centrin 3
|
|
chr3_-_127256215
|
3.580
|
NM_138593
|
Larp7
|
La ribonucleoprotein domain family, member 7
|
|
chr12_-_81213930
|
3.539
|
NM_007564
|
Zfp36l1
|
zinc finger protein 36, C3H type-like 1
|
|
chr9_+_51855254
|
3.500
|
NM_001104616
|
Rdx
|
radixin
|
|
chr7_+_3655602
|
3.482
|
NM_029767
|
Rps9
|
ribosomal protein S9
|
|
chr17_-_24664876
|
3.464
|
NM_001172113
NM_001172114
NM_153792
|
Traf7
|
TNF receptor-associated factor 7
|
|
chr2_+_172196161
|
3.440
|
|
Cstf1
|
cleavage stimulation factor, 3' pre-RNA, subunit 1
|
|
chr3_-_100773571
|
3.400
|
NM_001013026
|
Ttf2
|
transcription termination factor, RNA polymerase II
|
|
chr17_-_24664759
|
3.358
|
|
Traf7
|
TNF receptor-associated factor 7
|
|
chrX_+_160857716
|
3.335
|
NM_011081
|
Piga
|
phosphatidylinositol glycan anchor biosynthesis, class A
|
|
chr3_+_102799658
|
3.330
|
NM_025679
|
Sike1
|
suppressor of IKBKE 1
|
|
chrX_+_19941606
|
3.278
|
NM_133669
|
Rp2h
|
retinitis pigmentosa 2 homolog (human)
|
|
chr2_-_25080160
|
3.276
|
|
Tubb2c
|
tubulin, beta 2C
|
|
chr3_-_51364525
|
3.272
|
NM_080793
|
Setd7
|
SET domain containing (lysine methyltransferase) 7
|
|
chr15_+_102126686
|
3.190
|
NM_001014976
|
Espl1
|
extra spindle poles-like 1 (S. cerevisiae)
|
|
chr10_-_80962881
|
3.187
|
NM_010102
|
S1pr4
|
sphingosine-1-phosphate receptor 4
|
|
chr2_-_94252368
|
3.153
|
|
Api5
|
apoptosis inhibitor 5
|
|
chr11_+_116295406
|
3.098
|
NM_176902
|
Fam100b
|
family with sequence similarity 100, member B
|
|
chr17_-_25023253
|
3.027
|
NM_011956
|
Nubp2
|
nucleotide binding protein 2
|
|
chr1_+_58052814
|
3.010
|
NM_199007
|
Sgol2
|
shugoshin-like 2 (S. pombe)
|
|
chr6_+_125045998
|
2.944
|
NM_145979
|
Chd4
|
chromodomain helicase DNA binding protein 4
|
|
chr9_-_76893529
|
2.908
|
NM_012033
|
Tinag
|
tubulointerstitial nephritis antigen
|
|
chr9_+_56924152
|
2.891
|
NM_001110350
NM_011378
|
Sin3a
|
transcriptional regulator, SIN3A (yeast)
|
|
chr14_-_31942057
|
2.843
|
|
2010107H07Rik
|
RIKEN cDNA 2010107H07 gene
|
|
chr2_-_120229789
|
2.821
|
NM_001110496
NM_001110497
NM_173734
|
Tmem87a
|
transmembrane protein 87A
|
|
chr9_+_78463569
|
2.814
|
|
Cd109
|
CD109 antigen
|
|
chr7_+_3655919
|
2.804
|
|
Rps9
|
ribosomal protein S9
|
|
chr9_+_113840051
|
2.792
|
NM_001083319
NM_013699
|
Ubp1
|
upstream binding protein 1
|
|
chr7_-_29678773
|
2.707
|
|
Actn4
|
actinin alpha 4
|
|
chr13_-_119175893
|
2.684
|
NM_021556
|
Mrps30
|
mitochondrial ribosomal protein S30
|
|
chr15_-_83295010
|
2.680
|
NM_001159509
|
Pacsin2
|
protein kinase C and casein kinase substrate in neurons 2
|
|
chr10_-_10277885
|
2.661
|
NM_026405
|
Rab32
|
RAB32, member RAS oncogene family
|
|
chr2_-_155652217
|
2.661
|
|
Eif6
|
eukaryotic translation initiation factor 6
|
|
chr1_-_191511950
|
2.635
|
NM_001081363
|
Cenpf
|
centromere protein F
|
|
chr16_+_64851768
|
2.607
|
NM_178647
|
Cggbp1
|
CGG triplet repeat binding protein 1
|
|
chr11_-_70458401
|
2.597
|
|
Slc25a11
|
solute carrier family 25 (mitochondrial carrier oxoglutarate carrier), member 11
|
|
chr6_+_17699145
|
2.593
|
NM_001083315
NM_022332
|
St7
|
suppression of tumorigenicity 7
|
|
chr7_-_56136405
|
2.588
|
NM_001013368
|
E2f8
|
E2F transcription factor 8
|
|
chr11_-_72029093
|
2.586
|
NM_026068
|
Med31
|
mediator of RNA polymerase II transcription, subunit 31 homolog (yeast)
|
|
chr11_+_20443315
|
2.582
|
|
Sertad2
|
SERTA domain containing 2
|
|
chr8_-_64601861
|
2.530
|
NM_001136089
NM_011922
|
Anxa10
|
annexin A10
|
|
chr9_-_13631482
|
2.527
|
NM_026665
|
Cep57
|
centrosomal protein 57
|
|
chr9_-_119231439
|
2.490
|
|
Oxsr1
|
oxidative-stress responsive 1
|
|
chr7_+_49276094
|
2.451
|
NM_001111282
|
Hmg1l1
|
high-mobility group (nonhistone chromosomal) protein 1-like 1
|
|
chr6_-_98978185
|
2.444
|
NM_001197322
|
Foxp1
|
forkhead box P1
|
|
chr2_-_154429414
|
2.428
|
|
Pxmp4
|
peroxisomal membrane protein 4
|
|
chr7_-_134185928
|
2.413
|
NM_145588
|
Kif22
|
kinesin family member 22
|
|
chr5_-_121038602
|
2.412
|
NM_145853
|
Tpcn1
|
two pore channel 1
|
|
chr13_+_5860749
|
2.387
|
|
Klf6
|
Kruppel-like factor 6
|
|
chr3_+_37211467
|
2.378
|
NM_001008502
|
Bbs12
|
Bardet-Biedl syndrome 12 (human)
|
|
chr1_-_146622545
|
2.344
|
NM_022881
|
Rgs18
|
regulator of G-protein signaling 18
|
|
chr10_-_119549945
|
2.322
|
NM_080446
|
Helb
|
helicase (DNA) B
|
|
chr9_-_100446544
|
2.321
|
|
Nck1
|
non-catalytic region of tyrosine kinase adaptor protein 1
|
|
chr15_-_96529998
|
2.277
|
NM_175121
|
Slc38a2
|
solute carrier family 38, member 2
|
|
chr13_-_23837389
|
2.258
|
NM_175653
|
Hist1h3c
|
histone cluster 1, H3c
|
|
chr5_-_115608123
|
2.193
|
|
Mlec
|
malectin
|
|
chr7_+_82600419
|
2.161
|
NM_029332
|
Akap13
|
A kinase (PRKA) anchor protein 13
|
|
chr11_-_120434204
|
2.125
|
NM_011032
|
P4hb
|
prolyl 4-hydroxylase, beta polypeptide
|
|
chr15_-_83294927
|
2.118
|
|
Pacsin2
|
protein kinase C and casein kinase substrate in neurons 2
|
|
chr6_-_34267413
|
2.089
|
NM_009658
|
Akr1b3
|
aldo-keto reductase family 1, member B3 (aldose reductase)
|
|
chr13_-_72101161
|
2.083
|
NM_010573
|
Irx1
|
Iroquois related homeobox 1 (Drosophila)
|
|
chrX_+_151696991
|
2.054
|
NM_019736
|
Acot9
|
acyl-CoA thioesterase 9
|
|
chr7_-_56136367
|
2.053
|
|
E2f8
|
E2F transcription factor 8
|
|
chr13_+_21809496
|
2.038
|
NM_178206
|
Hist1h3h
|
histone cluster 1, H3h
|
|
chr7_-_134185894
|
2.037
|
|
Kif22
|
kinesin family member 22
|
|
chr11_-_99412144
|
2.029
|
NM_025524
|
Krtap3-3
|
keratin associated protein 3-3
|
|
chr8_+_55162885
|
2.024
|
NM_009506
|
Vegfc
|
vascular endothelial growth factor C
|
|
chr11_-_120434181
|
2.011
|
|
P4hb
|
prolyl 4-hydroxylase, beta polypeptide
|
|
chr2_+_102913831
|
2.008
|
NM_019735
|
Apip
|
APAF1 interacting protein
|
|
chr10_-_40021983
|
2.001
|
NM_007444
NM_009665
|
Amd2
Amd1
|
S-adenosylmethionine decarboxylase 2
S-adenosylmethionine decarboxylase 1
|
|
chr7_+_138175581
|
1.989
|
NM_007769
|
Dmbt1
|
deleted in malignant brain tumors 1
|
|
chr2_+_152673699
|
1.989
|
NM_001141975
NM_001141976
NM_001141977
NM_001141978
NM_028109
|
Tpx2
|
TPX2, microtubule-associated protein homolog (Xenopus laevis)
|
|
chr17_-_41071432
|
1.969
|
NM_031863
|
Cenpq
|
centromere protein Q
|