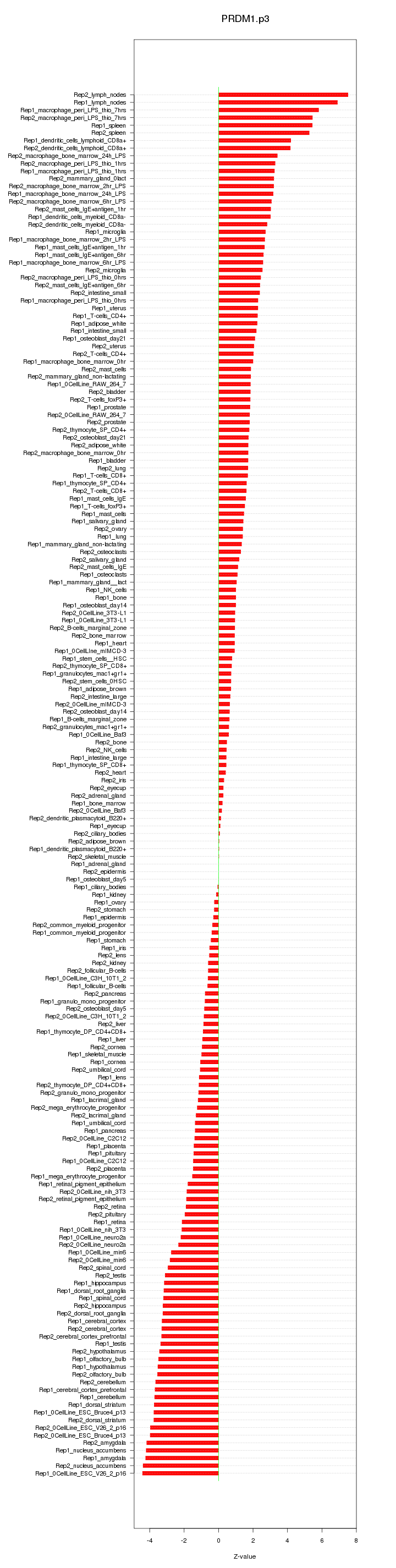
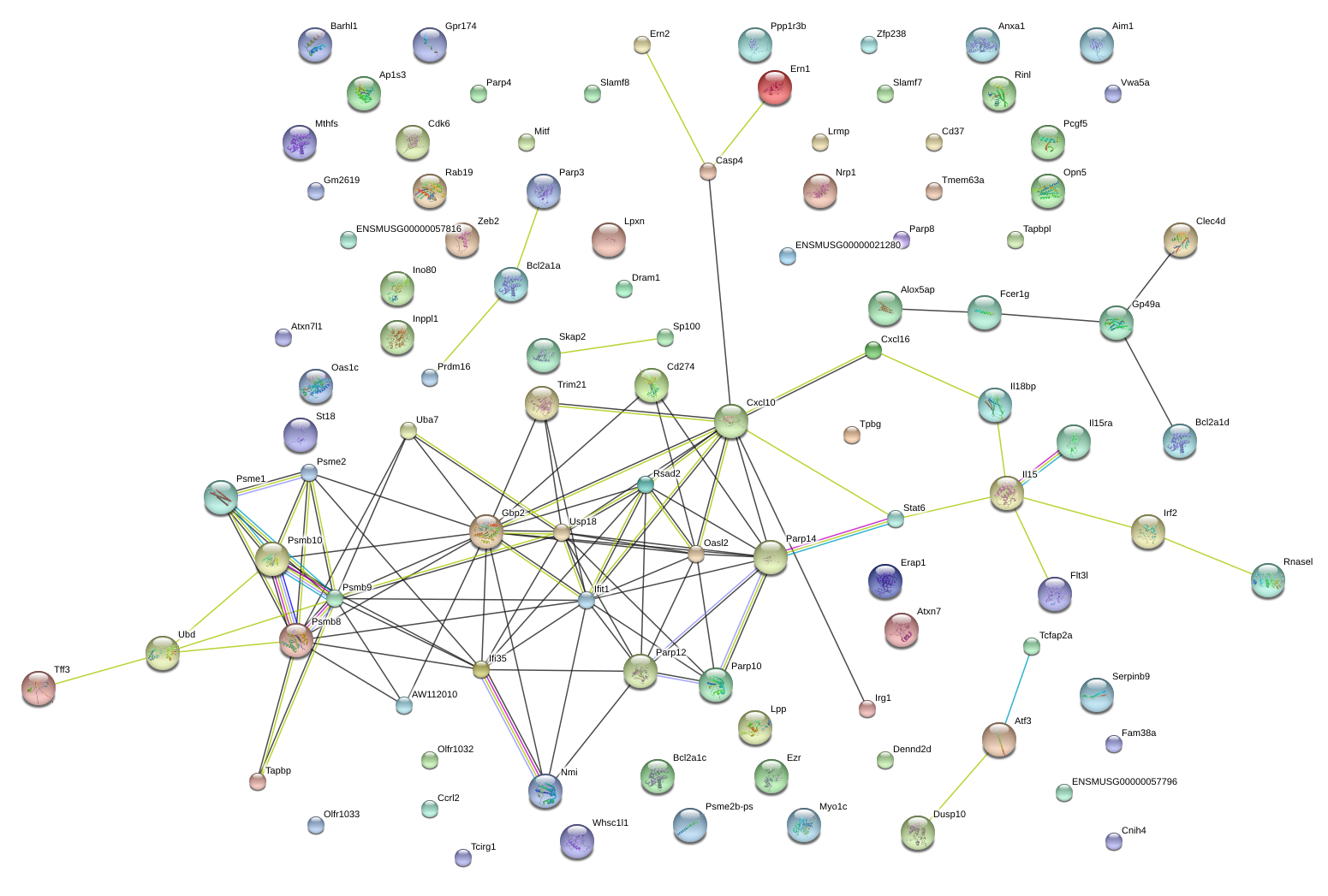
|
chr1_-_193007202
|
75.029
|
NM_007498
|
Atf3
|
activating transcription factor 3
|
|
chr9_+_5308846
|
67.477
|
NM_007609
|
Casp4
|
caspase 4, apoptosis-related cysteine peptidase
|
|
chr3_+_142283605
|
57.587
|
NM_010260
|
Gbp2
|
guanylate binding protein 2
|
|
chr19_+_34715368
|
56.882
|
NM_008331
|
Ifit1
|
interferon-induced protein with tetratricopeptide repeats 1
|
|
chr10_+_51210768
|
56.364
|
NM_013532
|
Lilrb4
|
leukocyte immunoglobulin-like receptor, subfamily B, member 4
|
|
chr7_-_52494453
|
53.344
|
|
Cd37
|
CD37 antigen
|
|
chr1_-_193006986
|
50.941
|
|
Atf3
|
activating transcription factor 3
|
|
chr19_+_29446941
|
49.737
|
|
Cd274
|
CD274 antigen
|
|
chr1_-_174520562
|
47.778
|
NM_029084
|
Slamf8
|
SLAM family member 8
|
|
chr6_+_145070255
|
43.876
|
NM_008511
|
Lrmp
|
lymphoid-restricted membrane protein
|
|
chr7_-_52494097
|
43.043
|
NM_007645
|
Cd37
|
CD37 antigen
|
|
chr8_-_84926440
|
42.169
|
NM_008357
|
Il15
|
interleukin 15
|
|
chr13_+_74777303
|
38.606
|
NM_030711
|
Erap1
|
endoplasmic reticulum aminopeptidase 1
|
|
chr2_-_28771940
|
37.862
|
NM_019446
NM_001164186
|
Barhl1
|
BarH-like 1 (Drosophila)
|
|
chr8_-_125075222
|
37.116
|
NM_001037298
|
Fam38a
|
family with sequence similarity 38, member A
|
|
chr3_+_106285556
|
36.984
|
|
Dennd2d
|
DENN/MADD domain containing 2D
|
|
chr6_-_39050297
|
35.828
|
|
Parp12
|
poly (ADP-ribose) polymerase family, member 12
|
|
chr1_-_79668530
|
35.139
|
NM_183027
|
Ap1s3
|
adaptor-related protein complex AP-1, sigma 3
|
|
chr19_-_11124897
|
32.833
|
NM_001177351
|
AW112010
|
expressed sequence AW112010
|
|
chr11_-_70273434
|
31.790
|
|
Cxcl16
|
chemokine (C-X-C motif) ligand 16
|
|
chr17_+_34335139
|
30.944
|
NM_010724
|
Psmb8
|
proteasome (prosome, macropain) subunit, beta type 8 (large multifunctional peptidase 7)
|
|
chr9_-_110959315
|
30.912
|
|
Ccrl2
|
chemokine (C-C motif) receptor-like 2
|
|
chr11_-_70273190
|
30.189
|
|
Cxcl16
|
chemokine (C-X-C motif) ligand 16
|
|
chr11_-_70273453
|
29.998
|
NM_023158
|
Cxcl16
|
chemokine (C-X-C motif) ligand 16
|
|
chr17_-_34324234
|
29.510
|
NM_013585
|
Psmb9
|
proteasome (prosome, macropain) subunit, beta type 9 (large multifunctional peptidase 2)
|
|
chr11_-_70273251
|
29.051
|
|
Cxcl16
|
chemokine (C-X-C motif) ligand 16
|
|
chr9_+_38525852
|
28.948
|
NM_001145957
NM_172767
|
Vwa5a
|
von Willebrand factor A domain containing 5A
|
|
chr12_-_27141303
|
28.945
|
NM_021384
|
Rsad2
|
radical S-adenosyl methionine domain containing 2
|
|
chr8_-_84927109
|
28.652
|
|
Il15
|
interleukin 15
|
|
chr11_-_70272909
|
28.593
|
|
Cxcl16
|
chemokine (C-X-C motif) ligand 16
|
|
chr19_-_3905229
|
28.455
|
NM_016921
|
Tcirg1
|
T-cell, immune regulator 1, ATPase, H+ transporting, lysosomal V0 protein A3
|
|
chr17_+_34056555
|
27.611
|
|
Tapbp
|
TAP binding protein
|
|
chr15_-_76073789
|
27.596
|
|
Parp10
|
poly (ADP-ribose) polymerase family, member 10
|
|
chr9_-_106378494
|
27.282
|
|
Parp3
|
poly (ADP-ribose) polymerase family, member 3
|
|
chr9_+_107877887
|
27.036
|
NM_023738
|
Uba7
|
ubiquitin-like modifier activating enzyme 7
|
|
chr5_-_92776027
|
26.927
|
|
Cxcl10
|
chemokine (C-X-C motif) ligand 10
|
|
chr7_-_109713935
|
26.789
|
NM_001082552
NM_009277
|
Trim21
|
tripartite motif-containing 21
|
|
chr13_+_33096472
|
26.547
|
|
Serpinb9
|
serine (or cysteine) peptidase inhibitor, clade B, member 9
|
|
chr17_+_34056585
|
26.244
|
|
Tapbp
|
TAP binding protein
|
|
chr11_-_70273333
|
25.989
|
|
Cxcl16
|
chemokine (C-X-C motif) ligand 16
|
|
chr15_-_76073869
|
25.953
|
NM_001163575
NM_001163576
|
Parp10
|
poly (ADP-ribose) polymerase family, member 10
|
|
chr15_-_76073854
|
25.623
|
|
Parp10
|
poly (ADP-ribose) polymerase family, member 10
|
|
chr10_-_87819740
|
25.282
|
|
Dram1
|
DNA-damage regulated autophagy modulator 1
|
|
chr11_-_106257752
|
25.160
|
|
Ern1
|
endoplasmic reticulum (ER) to nucleus signalling 1
|
|
chr12_-_27141277
|
24.677
|
|
Rsad2
|
radical S-adenosyl methionine domain containing 2
|
|
chr15_-_76073582
|
23.869
|
|
Parp10
|
poly (ADP-ribose) polymerase family, member 10
|
|
chr17_+_34056798
|
23.758
|
|
Tapbp
|
TAP binding protein
|
|
chr8_-_108461909
|
23.631
|
|
Psmb10
|
proteasome (prosome, macropain) subunit, beta type 10
|
|
chr9_-_88626613
|
23.346
|
NM_007536
|
Bcl2a1d
|
B-cell leukemia/lymphoma 2 related protein A1d
|
|
chr2_+_85860710
|
23.014
|
NM_146578
|
Olfr1033
|
olfactory receptor 1033
|
|
chr3_+_106285348
|
22.748
|
NM_001093754
|
Dennd2d
|
DENN/MADD domain containing 2D
|
|
chr9_-_106378901
|
21.982
|
NM_145619
|
Parp3
|
poly (ADP-ribose) polymerase family, member 3
|
|
chr8_-_108462291
|
21.932
|
NM_013640
|
Psmb10
|
proteasome (prosome, macropain) subunit, beta type 10
|
|
chr19_-_20465115
|
21.819
|
|
Anxa1
|
annexin A1
|
|
chr14_+_103446209
|
21.560
|
NM_008392
|
Irg1
|
immunoresponsive gene 1
|
|
chr12_-_27141171
|
21.334
|
|
Rsad2
|
radical S-adenosyl methionine domain containing 2
|
|
chr13_+_33096402
|
19.488
|
NM_009256
|
Serpinb9
|
serine (or cysteine) peptidase inhibitor, clade B, member 9
|
|
chrX_+_104479684
|
19.475
|
NM_001177782
|
Gpr174
|
G protein-coupled receptor 174
|
|
chr19_+_12873115
|
19.359
|
|
Lpxn
|
leupaxin
|
|
chr14_+_57194455
|
19.192
|
NM_001145978
|
Parp4
|
poly (ADP-ribose) polymerase family, member 4
|
|
chr2_-_51828440
|
18.801
|
|
Nmi
|
N-myc (and STAT) interactor
|
|
chr14_-_56209708
|
18.782
|
NM_011190
|
Psme2
Psme2b-ps
|
proteasome (prosome, macropain) 28 subunit, beta
protease (prosome, macropain) 28 subunit beta B, pseudogene
|
|
chr6_-_51962458
|
18.621
|
NM_018773
|
Skap2
|
src family associated phosphoprotein 2
|
|
chr17_+_34056367
|
17.708
|
NM_001025313
NM_009318
|
Tapbp
|
TAP binding protein
|
|
chr17_-_6943173
|
17.641
|
|
Ezr
|
ezrin
|
|
chr12_+_112655640
|
17.435
|
NM_028807
|
1200009I06Rik
|
RIKEN cDNA 1200009I06 gene
|
|
chr11_+_101309725
|
17.031
|
NM_027320
|
Ifi35
|
interferon-induced protein 35
|
|
chr6_+_97879810
|
16.971
|
NM_001178049
|
Mitf
|
microphthalmia-associated transcription factor
|
|
chr16_+_24393435
|
16.815
|
NM_178665
|
Lpp
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr1_+_87546558
|
16.587
|
NM_013673
|
Sp100
|
nuclear antigen Sp100
|
|
chr12_+_33987379
|
16.583
|
NM_001033436
|
Atxn7l1
|
ataxin 7-like 1
|
|
chr10_+_127080013
|
16.028
|
NM_009284
|
Stat6
|
signal transducer and activator of transcription 6
|
|
chr9_+_88851757
|
16.009
|
NM_009742
|
Bcl2a1a
|
B-cell leukemia/lymphoma 2 related protein A1a
|
|
chr14_+_56197282
|
15.892
|
NM_011189
|
Psme1
|
proteasome (prosome, macropain) 28 subunit, alpha
|
|
chr6_+_123212124
|
15.462
|
NM_001163161
NM_010819
|
Clec4d
|
C-type lectin domain family 4, member d
|
|
chr8_+_131028275
|
14.944
|
|
Nrp1
|
neuropilin 1
|
|
chr5_-_92776160
|
14.155
|
|
Cxcl10
|
chemokine (C-X-C motif) ligand 10
|
|
chr10_+_127080251
|
13.938
|
|
Stat6
|
signal transducer and activator of transcription 6
|
|
chr1_-_173164404
|
13.719
|
NM_010185
|
Fcer1g
|
Fc receptor, IgE, high affinity I, gamma polypeptide
|
|
chr16_-_35871459
|
13.697
|
NM_001039530
|
Parp14
|
poly (ADP-ribose) polymerase family, member 14
|
|
chr6_+_39331379
|
13.143
|
NM_011226
|
Rab19
|
RAB19, member RAS oncogene family
|
|
chr7_+_29573987
|
12.881
|
NM_177158
|
Rinl
|
Ras and Rab interactor-like
|
|
chr5_+_150076605
|
12.303
|
NM_009663
|
Alox5ap
|
arachidonate 5-lipoxygenase activating protein
|
|
chr6_+_123212210
|
12.243
|
|
Clec4d
|
C-type lectin domain family 4, member d
|
|
chr1_+_182872588
|
12.115
|
NM_144794
|
Tmem63a
|
transmembrane protein 63a
|
|
chr1_+_6720153
|
11.564
|
|
St18
|
suppression of tumorigenicity 18
|
|
chr7_-_52391419
|
11.265
|
|
Flt3l
|
FMS-like tyrosine kinase 3 ligand
|
|
chr19_-_5845001
|
11.136
|
|
Neat1
|
nuclear paraspeckle assembly transcript 1 (non-protein coding)
|
|
chr9_-_106378454
|
11.067
|
|
Parp3
|
poly (ADP-ribose) polymerase family, member 3
|
|
chr1_+_6720087
|
10.757
|
NM_173868
|
St18
|
suppression of tumorigenicity 18
|
|
chr9_-_88626434
|
10.637
|
|
Bcl2a1d
|
B-cell leukemia/lymphoma 2 related protein A1d
|
|
chr2_-_119303348
|
10.515
|
NM_026574
|
Ino80
|
INO80 homolog (S. cerevisiae)
|
|
chr13_-_40825789
|
10.261
|
NM_001122948
|
Tcfap2a
|
transcription factor AP-2, alpha
|
|
chr1_+_185858324
|
10.255
|
NM_022019
|
Dusp10
|
dual specificity phosphatase 10
|
|
chr13_-_117814322
|
9.908
|
NM_001081009
|
Parp8
|
poly (ADP-ribose) polymerase family, member 8
|
|
chr7_-_108986589
|
9.902
|
NM_010567
|
Inppl1
|
inositol polyphosphate phosphatase-like 1
|
|
chr17_+_37330836
|
9.783
|
NM_023137
|
Ubd
|
ubiquitin D
|
|
chr6_-_125181731
|
9.297
|
NM_145391
|
Tapbpl
|
TAP binding protein-like
|
|
chr2_-_44965866
|
9.224
|
|
Zeb2
|
zinc finger E-box binding homeobox 2
|
|
chr8_+_47825098
|
9.033
|
NM_008391
|
Irf2
|
interferon regulatory factor 2
|
|
chr7_-_52391756
|
8.792
|
NM_013520
|
Flt3l
|
FMS-like tyrosine kinase 3 ligand
|
|
chr5_+_115347505
|
8.153
|
|
Oasl2
|
2'-5' oligoadenylate synthetase-like 2
|
|
chr19_+_12873063
|
7.742
|
NM_134152
|
Lpxn
|
leupaxin
|
|
chr1_+_183081066
|
7.691
|
|
Cnih4
|
cornichon homolog 4 (Drosophila)
|
|
chr1_+_155596555
|
7.636
|
NM_011882
|
Rnasel
|
ribonuclease L (2', 5'-oligoisoadenylate synthetase-dependent)
|
|
chr8_+_36438761
|
7.632
|
NM_177741
|
Ppp1r3b
|
protein phosphatase 1, regulatory (inhibitor) subunit 3B
|
|
chr7_-_109166668
|
7.557
|
NM_010531
|
Il18bp
|
interleukin 18 binding protein
|
|
chr4_-_154010868
|
7.259
|
NM_001177995
NM_027504
|
Prdm16
|
PR domain containing 16
|
|
chr19_+_36484237
|
7.240
|
|
Pcgf5
|
polycomb group ring finger 5
|
|
chr5_+_3343861
|
7.235
|
|
Cdk6
|
cyclin-dependent kinase 6
|
|
chr9_-_110959702
|
7.234
|
NM_017466
|
Ccrl2
|
chemokine (C-C motif) receptor-like 2
|
|
chr2_+_11627013
|
7.217
|
NM_133836
|
Il15ra
|
interleukin 15 receptor, alpha chain
|
|
chr1_-_173579128
|
7.075
|
|
Slamf7
|
SLAM family member 7
|
|
chr8_+_26750755
|
6.889
|
NM_001081269
|
Whsc1l1
|
Wolf-Hirschhorn syndrome candidate 1-like 1 (human)
|
|
chr6_+_121195885
|
6.508
|
NM_011909
|
Usp18
|
ubiquitin specific peptidase 18
|
|
chr1_+_179372452
|
6.447
|
|
Zfp238
|
zinc finger protein 238
|
|
chr14_+_14845004
|
6.377
|
NM_139227
|
Atxn7
|
ataxin 7
|
|
chr9_+_5298516
|
6.225
|
NM_009807
|
Casp1
|
caspase 1
|
|
chr8_-_85856284
|
6.147
|
NM_001170691
NM_178736
|
Elmod2
|
ELMO domain containing 2
|
|
chr5_+_28492235
|
6.144
|
NM_010134
|
En2
|
engrailed 2
|
|
chr6_-_39068241
|
6.137
|
|
Parp12
|
poly (ADP-ribose) polymerase family, member 12
|
|
chr13_-_3917356
|
6.118
|
NM_019671
|
Net1
|
neuroepithelial cell transforming gene 1
|
|
chr5_-_105668551
|
5.997
|
NM_001039646
|
Gbp10
|
guanylate-binding protein 10
|
|
chr7_-_109166133
|
5.877
|
|
Il18bp
|
interleukin 18 binding protein
|
|
chr2_-_25221101
|
5.761
|
NM_001033293
|
Uap1l1
|
UDP-N-acteylglucosamine pyrophosphorylase 1-like 1
|
|
chr15_-_77277889
|
5.749
|
NM_001024848
|
Apol7b
|
apolipoprotein L 7b
|
|
chr10_-_87819765
|
5.717
|
NM_027878
|
Dram1
|
DNA-damage regulated autophagy modulator 1
|
|
chr5_-_105411580
|
5.701
|
NM_030239
|
Abcg3
|
ATP-binding cassette, sub-family G (WHITE), member 3
|
|
chr17_-_35309483
|
5.659
|
|
Aif1
|
allograft inflammatory factor 1
|
|
chr19_-_5845462
|
5.526
|
|
Neat1
|
nuclear paraspeckle assembly transcript 1 (non-protein coding)
|
|
chr11_+_6460185
|
5.326
|
NM_001190344
|
Ccm2
|
cerebral cavernous malformation 2 homolog (human)
|
|
chr3_-_115710131
|
5.295
|
NM_023214
|
Slc30a7
|
solute carrier family 30 (zinc transporter), member 7
|
|
chr2_+_11627385
|
5.238
|
NM_008358
|
Il15ra
|
interleukin 15 receptor, alpha chain
|
|
chr8_+_95351212
|
5.113
|
NM_008610
|
Mmp2
|
matrix metallopeptidase 2
|
|
chr4_+_15211137
|
5.109
|
|
Tmem64
|
transmembrane protein 64
|
|
chr4_-_149377238
|
5.064
|
|
H6pd
|
hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase)
|
|
chr3_-_142054771
|
5.034
|
NM_001190857
NM_022554
|
Pdlim5
|
PDZ and LIM domain 5
|
|
chr13_-_117814079
|
4.957
|
|
Parp8
|
poly (ADP-ribose) polymerase family, member 8
|
|
chr9_+_108417075
|
4.721
|
|
Qars
|
glutaminyl-tRNA synthetase
|
|
chr17_+_35187325
|
4.688
|
|
Clic1
|
chloride intracellular channel 1
|
|
chr16_-_97684510
|
4.601
|
NM_010846
|
Mx1
|
myxovirus (influenza virus) resistance 1
|
|
chr5_+_115373333
|
4.582
|
|
Oasl1
|
2'-5' oligoadenylate synthetase-like 1
|
|
chr17_-_36282006
|
4.522
|
|
H2-D1
|
histocompatibility 2, D region locus 1
|
|
chr13_-_101017247
|
4.487
|
NM_010870
|
Naip5
|
NLR family, apoptosis inhibitory protein 5
|
|
chr5_-_141476549
|
4.376
|
NM_175362
|
Card11
|
caspase recruitment domain family, member 11
|
|
chr7_-_52391649
|
4.250
|
|
Flt3l
|
FMS-like tyrosine kinase 3 ligand
|
|
chr8_-_46495663
|
4.223
|
|
Tlr3
|
toll-like receptor 3
|
|
chr11_-_120434204
|
4.132
|
NM_011032
|
P4hb
|
prolyl 4-hydroxylase, beta polypeptide
|
|
chr2_-_132078946
|
4.027
|
|
Pcna
|
proliferating cell nuclear antigen
|
|
chr18_-_42110847
|
4.023
|
NM_029942
|
Prelid2
|
PRELI domain containing 2
|
|
chr12_+_111685516
|
4.018
|
|
Ppp2r5c
|
protein phosphatase 2, regulatory subunit B (B56), gamma isoform
|
|
chr10_+_127759512
|
4.000
|
NM_019953
|
Cnpy2
|
canopy 2 homolog (zebrafish)
|
|
chr7_-_148354562
|
3.921
|
NM_001172101
NM_145135
|
Rnh1
|
ribonuclease/angiogenin inhibitor 1
|
|
chr13_-_100972022
|
3.870
|
NM_001126182
NM_010872
|
Naip2
|
NLR family, apoptosis inhibitory protein 2
|
|
chr10_-_29255606
|
3.794
|
NM_028351
|
Rspo3
|
R-spondin 3 homolog (Xenopus laevis)
|
|
chr9_+_108410335
|
3.720
|
NM_133794
|
Qars
|
glutaminyl-tRNA synthetase
|
|
chr8_-_46495958
|
3.717
|
|
Tlr3
|
toll-like receptor 3
|
|
chr13_+_34254827
|
3.681
|
NM_001101430
NM_001110515
|
Psmg4
|
proteasome (prosome, macropain) assembly chaperone 4
|
|
chr8_-_46495890
|
3.667
|
NM_126166
|
Tlr3
|
toll-like receptor 3
|
|
chr5_-_105482574
|
3.575
|
NM_029509
|
Gbp8
|
guanylate-binding protein 8
|
|
chr4_+_86394436
|
3.567
|
NM_001081014
NM_184088
|
Dennd4c
|
DENN/MADD domain containing 4C
|
|
chr4_-_118954780
|
3.540
|
|
Ybx1
|
Y box protein 1
|
|
chr2_+_4480799
|
3.438
|
NM_001177844
|
Frmd4a
|
FERM domain containing 4A
|
|
chr13_-_31065909
|
3.370
|
NM_025588
|
Exoc2
|
exocyst complex component 2
|
|
chr11_-_99016390
|
3.333
|
NM_007719
|
Ccr7
|
chemokine (C-C motif) receptor 7
|
|
chr2_+_32391249
|
3.306
|
|
Fam102a
|
family with sequence similarity 102, member A
|
|
chr18_+_83084023
|
3.136
|
NM_001177464
|
Zfp516
|
zinc finger protein 516
|
|
chr6_+_34813131
|
3.026
|
NM_197986
|
Tmem140
|
transmembrane protein 140
|
|
chr7_+_97535516
|
3.006
|
|
Sytl2
|
synaptotagmin-like 2
|
|
chr11_+_4167080
|
3.006
|
NM_001039537
|
Lif
|
leukemia inhibitory factor
|
|
chr10_+_127759640
|
2.974
|
|
Cnpy2
|
canopy 2 homolog (zebrafish)
|
|
chr3_-_58905678
|
2.840
|
NM_173398
|
Gpr171
|
G protein-coupled receptor 171
|
|
chr13_+_34254902
|
2.794
|
|
Psmg4
|
proteasome (prosome, macropain) assembly chaperone 4
|
|
chr18_+_89366818
|
2.737
|
NM_001039149
NM_178687
|
Cd226
|
CD226 antigen
|
|
chr2_+_27741864
|
2.698
|
NM_015734
|
Col5a1
|
collagen, type V, alpha 1
|
|
chr18_-_52689504
|
2.602
|
|
Lox
|
lysyl oxidase
|
|
chr12_-_86644527
|
2.544
|
|
Nek9
|
NIMA (never in mitosis gene a)-related expressed kinase 9
|
|
chr2_-_132078887
|
2.530
|
NM_011045
|
Pcna
|
proliferating cell nuclear antigen
|
|
chr7_+_97535490
|
2.443
|
NM_001040087
NM_001040088
|
Sytl2
|
synaptotagmin-like 2
|
|
chr1_-_185857753
|
2.409
|
NM_028516
|
1700056E22Rik
|
RIKEN cDNA 1700056E22 gene
|
|
chr14_+_56188462
|
2.319
|
|
|
|
|
chr11_-_61058671
|
2.310
|
|
|
|
|
chr13_-_101087625
|
2.303
|
NM_021545
|
Naip6
Naip7
|
NLR family, apoptosis inhibitory protein 6
NLR family, apoptosis inhibitory protein 7
|
|
chr10_+_117878101
|
2.265
|
NM_008337
|
Ifng
|
interferon gamma
|
|
chr15_+_77307476
|
2.241
|
NM_177744
|
Apol10a
|
apolipoprotein L 10a
|
|
chr11_-_120434075
|
2.206
|
|
P4hb
|
prolyl 4-hydroxylase, beta polypeptide
|
|
chr6_-_71582562
|
2.163
|
NM_173001
|
Kdm3a
|
lysine (K)-specific demethylase 3A
|
|
chr4_-_124359695
|
2.120
|
|
Utp11l
|
UTP11-like, U3 small nucleolar ribonucleoprotein, (yeast)
|
|
chr9_+_35024031
|
2.078
|
|
Srpr
|
signal recognition particle receptor ('docking protein')
|
|
chr2_-_51790406
|
2.044
|
NM_001141981
NM_030243
|
Rbm43
|
RNA binding motif protein 43
|
|
chr2_+_145648290
|
2.032
|
|
Rin2
|
Ras and Rab interactor 2
|
|
chr15_+_25772997
|
2.027
|
NM_001034851
|
Fam134b
|
family with sequence similarity 134, member B
|
|
chr9_-_50425561
|
1.976
|
NM_001134902
NM_212449
|
AU019823
|
expressed sequence AU019823
|
|
chr1_+_45401659
|
1.960
|
|
Col3a1
|
collagen, type III, alpha 1
|
|
chr9_+_114239252
|
1.953
|
NM_007535
|
Bcl2a1c
|
B-cell leukemia/lymphoma 2 related protein A1c
|
|
chr17_+_30763879
|
1.947
|
NM_013811
|
Dnahc8
|
dynein, axonemal, heavy chain 8
|
|
chr10_-_36855292
|
1.946
|
|
Marcks
|
myristoylated alanine rich protein kinase C substrate
|
|
chr3_-_9833502
|
1.943
|
NM_001195031
NM_053182
|
Pag1
|
phosphoprotein associated with glycosphingolipid microdomains 1
|
|
chr9_-_37063214
|
1.939
|
NM_001145960
NM_020258
|
Slc37a2
|
solute carrier family 37 (glycerol-3-phosphate transporter), member 2
|
|
chr8_+_91220959
|
1.894
|
|
Cyld
|
cylindromatosis (turban tumor syndrome)
|