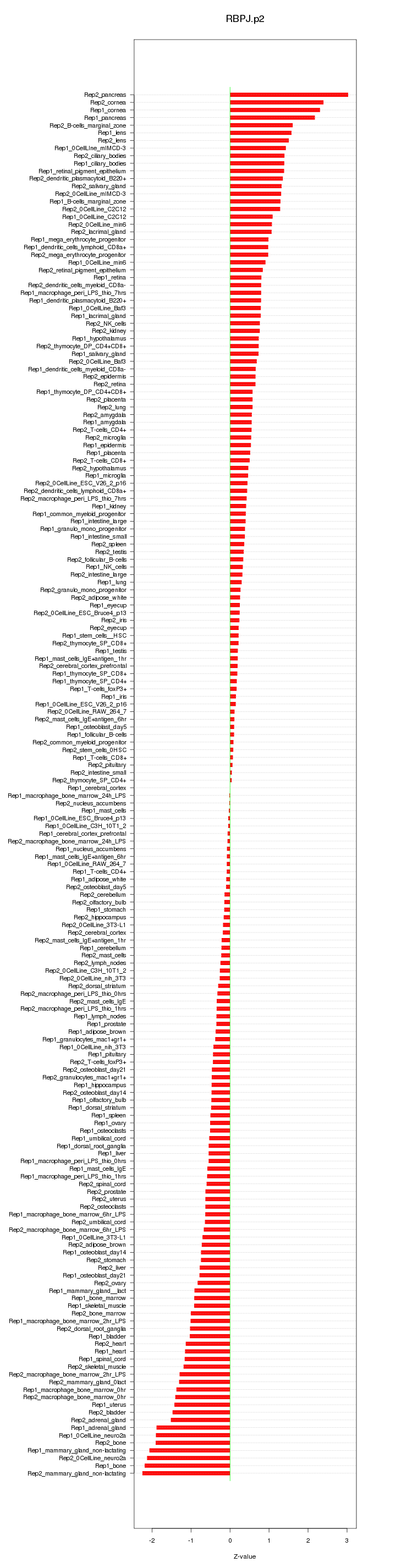
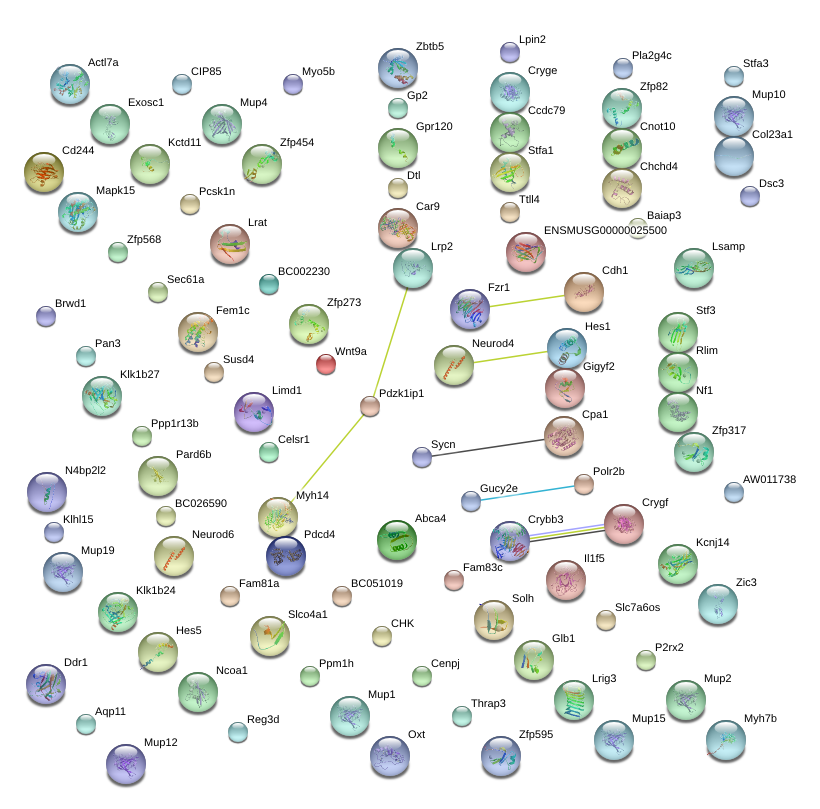
|
chr9_-_69989168
|
8.984
|
|
Fam81a
|
family with sequence similarity 81, member A
|
|
chr9_-_69989285
|
8.216
|
NM_029784
|
Fam81a
|
family with sequence similarity 81, member A
|
|
chr2_+_180195682
|
6.406
|
NM_148933
|
Slco4a1
|
solute carrier organic anion transporter family, member 4a1
|
|
chr4_+_154335029
|
6.343
|
NM_010419
|
Hes5
|
hairy and enhancer of split 5 (Drosophila)
|
|
chr7_-_126602780
|
5.611
|
NM_025989
|
Gp2
|
glycoprotein 2 (zymogen granule membrane)
|
|
chr5_-_113510602
|
4.341
|
NM_001159650
NM_021352
|
Crybb3
|
crystallin, beta B3
|
|
chr2_-_69424085
|
4.337
|
NM_001081088
|
Lrp2
|
low density lipoprotein receptor-related protein 2
|
|
chr6_-_91423388
|
4.163
|
NM_133928
|
Chchd4
|
coiled-coil-helix-coiled-coil-helix domain containing 4
|
|
chr7_+_29325883
|
4.084
|
NM_026716
|
Sycn
|
syncollin
|
|
chr10_+_122116066
|
4.010
|
|
Ppm1h
|
protein phosphatase 1H (PP2C domain containing)
|
|
chr19_+_53966720
|
3.761
|
NM_011050
|
Pdcd4
|
programmed cell death 4
|
|
chr1_-_65097587
|
3.637
|
NM_007777
|
Cryge
|
crystallin, gamma E
|
|
chr15_-_85864206
|
3.583
|
NM_009886
|
Celsr1
|
cadherin, EGF LAG seven-pass G-type receptor 1 (flamingo homolog, Drosophila)
|
|
chr17_-_25393220
|
3.541
|
NM_001163270
|
Baiap3
|
BAI1-associated protein 3
|
|
chr15_+_75824198
|
3.436
|
NM_177922
|
Mapk15
|
mitogen-activated protein kinase 15
|
|
chr9_+_19426528
|
3.401
|
NM_172918
|
Zfp317
|
zinc finger protein 317
|
|
chr1_+_74726341
|
3.323
|
|
Ttll4
|
tubulin tyrosine ligase-like family, member 4
|
|
chr6_+_30589217
|
3.268
|
NM_025350
|
Cpa1
|
carboxypeptidase A1
|
|
chr1_+_89223558
|
3.118
|
NM_146112
NM_001110212
|
Gigyf2
|
GRB10 interacting GYF protein 2
|
|
chr6_+_30589440
|
2.912
|
|
Cpa1
|
carboxypeptidase A1
|
|
chr11_-_69050508
|
2.857
|
NM_008192
|
Gucy2e
|
guanylate cyclase 2e
|
|
chr4_+_155577378
|
2.857
|
|
AW011738
|
expressed sequence AW011738
|
|
chr7_+_51443605
|
2.850
|
NM_010643
|
Klk1b24
|
kallikrein 1-related peptidase b24
|
|
chr8_-_107033781
|
2.802
|
NM_180958
|
Ccdc79
|
coiled-coil domain containing 79
|
|
chr5_-_110772051
|
2.771
|
NM_001164833
NM_001164834
NM_153400
|
P2rx2
|
purinergic receptor P2X, ligand-gated ion channel, 2
|
|
chrX_+_91480268
|
2.742
|
NM_001039059
NM_001039060
NM_001039061
NM_153165
|
Klhl15
|
kelch-like 15 (Drosophila)
|
|
chr4_-_59973470
|
2.739
|
NM_008648
|
Mup4
|
major urinary protein 4
|
|
chr6_-_78328836
|
2.692
|
NM_001161741
NM_013893
|
Reg3d
|
regenerating islet-derived 3 delta
|
|
chr7_-_30857837
|
2.689
|
NM_177889
|
Zfp82
|
zinc finger protein 82
|
|
chr4_-_106850812
|
2.632
|
NM_025617
|
2210012G02Rik
|
RIKEN cDNA 2210012G02 gene
|
|
chr2_+_167906462
|
2.624
|
NM_021409
|
Pard6b
|
par-6 (partitioning defective 6) homolog beta (C. elegans)
|
|
chr16_+_30065419
|
2.572
|
NM_008235
|
Hes1
|
hairy and enhancer of split 1 (Drosophila)
|
|
chr11_-_69694379
|
2.571
|
NM_153143
|
Kctd11
|
potassium channel tetramerisation domain containing 11
|
|
chr7_-_51926142
|
2.487
|
|
Myh14
|
myosin, heavy polypeptide 14
|
|
chr18_-_20160536
|
2.417
|
NM_007882
|
Dsc3
|
desmocollin 3
|
|
chr2_+_24132788
|
2.350
|
NM_019451
|
Il1f5
|
interleukin 1 family, member 5 (delta)
|
|
chr12_-_113146259
|
2.306
|
NM_011625
|
Ppp1r13b
|
protein phosphatase 1, regulatory (inhibitor) subunit 13B
|
|
chr2_+_130401908
|
2.232
|
NM_011025
|
Oxt
|
oxytocin
|
|
chr13_+_67914748
|
2.228
|
NM_198322
|
Zfp273
|
zinc finger protein 273
|
|
chrX_-_101176600
|
2.228
|
NM_011276
|
Rlim
|
ring finger protein, LIM domain interacting
|
|
chr17_-_35837432
|
2.202
|
NM_001198833
|
Ddr1
|
discoidin domain receptor family, member 1
|
|
chrX_+_55283804
|
2.196
|
NM_009575
|
Zic3
|
zinc finger protein of the cerebellum 3
|
|
chr7_+_13512764
|
2.165
|
NM_029809
|
2310014L17Rik
|
RIKEN cDNA 2310014L17 gene
|
|
chr1_+_184695026
|
2.153
|
NM_144796
|
Susd4
|
sushi domain containing 4
|
|
chr14_+_57430806
|
2.146
|
|
2410022M11Rik
|
RIKEN cDNA 2410022M11 gene
|
|
chr1_+_173489323
|
2.118
|
NM_018729
|
Cd244
LOC677008
|
CD244 natural killer cell receptor 2B4
natural killer cell receptor 2B4-like
|
|
chr11_+_51103421
|
2.116
|
NM_153393
|
Col23a1
|
collagen, type XXIII, alpha 1
|
|
chr3_+_121747377
|
2.101
|
NM_007378
|
Abca4
|
ATP-binding cassette, sub-family A (ABC1), member 4
|
|
chr4_+_43519895
|
2.092
|
NM_139305
|
Car9
|
carbonic anhydrase 9
|
|
chr7_+_51307659
|
2.087
|
NM_020268
|
Klk1b27
|
kallikrein 1-related peptidase b27
|
|
chr4_-_45024999
|
2.085
|
NM_001163283
NM_001163284
NM_173399
|
Zbtb5
|
zinc finger and BTB domain containing 5
|
|
chr17_-_26122588
|
2.065
|
|
Solh
|
small optic lobes homolog (Drosophila)
|
|
chr7_-_104886756
|
2.052
|
NM_175105
|
Aqp11
|
aquaporin 11
|
|
chr11_-_50700944
|
1.995
|
NM_172794
|
Zfp454
|
zinc finger protein 454
|
|
chr2_-_155660587
|
1.969
|
NM_027788
|
Fam83c
|
family with sequence similarity 83, member C
|
|
chr9_-_114549148
|
1.951
|
NM_153585
|
Cnot10
|
CCR4-NOT transcription complex, subunit 10
|
|
chr9_-_114549261
|
1.933
|
|
Cnot10
|
CCR4-NOT transcription complex, subunit 10
|
|
chr13_-_67433435
|
1.931
|
NM_177622
|
Zfp595
|
zinc finger protein 595
|
|
chr10_+_125403274
|
1.906
|
NM_177152
|
Lrig3
|
leucine-rich repeats and immunoglobulin-like domains 3
|
|
chr4_+_56815248
|
1.900
|
|
BC026590
|
cDNA sequence BC026590
|
|
chr16_+_36277233
|
1.882
|
NM_001082543
|
Stfa1
|
stefin A1
|
|
chr7_-_53080056
|
1.858
|
NM_145963
|
Kcnj14
|
potassium inwardly-rectifying channel, subfamily J, member 14
|
|
chr11_+_79153388
|
1.856
|
NM_010897
|
Nf1
|
neurofibromatosis 1
|
|
chr7_+_30768973
|
1.843
|
NM_001033355
NM_001167872
NM_001167873
|
Zfp568
|
zinc finger protein 568
|
|
chr4_-_125879832
|
1.840
|
NM_146153
|
Thrap3
|
thyroid hormone receptor associated protein 3
|
|
chr12_-_101397818
|
1.825
|
NM_183155
|
BC002230
|
cDNA sequence BC002230
|
|
chr4_+_56756283
|
1.808
|
NM_009611
|
Actl7a
|
actin-like 7a
|
|
chr16_-_96304116
|
1.785
|
|
Brwd1
|
bromodomain and WD repeat domain containing 1
|
|
chr7_+_4972164
|
1.774
|
|
Zfp865
|
zinc finger protein 865
|
|
chr11_+_59120429
|
1.758
|
NM_139298
|
Wnt9a
|
wingless-type MMTV integration site 9A
|
|
chr12_-_4483975
|
1.751
|
|
Ncoa1
|
nuclear receptor coactivator 1
|
|
chr7_-_148399978
|
1.750
|
NM_028050
|
1600016N20Rik
|
RIKEN cDNA 1600016N20 gene
|
|
chr17_-_26122737
|
1.743
|
|
Solh
|
small optic lobes homolog (Drosophila)
|
|
chr16_+_41533317
|
1.733
|
NM_175548
|
Lsamp
|
limbic system-associated membrane protein
|
|
chr18_-_46685557
|
1.733
|
NM_173423
|
Fem1c
|
fem-1 homolog c (C.elegans)
|
|
chr4_+_114761312
|
1.729
|
NM_001164557
NM_001164558
NM_026018
|
Pdzk1ip1
|
PDZK1 interacting protein 1
|
|
chr13_-_67585160
|
1.712
|
NM_001076791
|
Zfp874b
|
zinc finger protein 874b
|
|
chr19_-_42007761
|
1.703
|
NM_001164561
NM_025644
|
Exosc1
|
exosome component 1
|
|
chr17_+_71533317
|
1.696
|
NM_022882
|
Lpin2
|
lipin 2
|
|
chr8_+_109127245
|
1.695
|
NM_009864
|
Cdh1
|
cadherin 1
|
|
chr5_+_148241736
|
1.690
|
|
Pan3
|
PAN3 polyA specific ribonuclease subunit homolog (S. cerevisiae)
|
|
chr9_+_123387818
|
1.689
|
NM_013860
|
Limd1
|
LIM domains containing 1
|
|
chr8_-_108734817
|
1.662
|
NM_001007567
|
Slc7a6os
|
solute carrier family 7, member 6 opposite strand
|
|
chr10_-_129717244
|
1.655
|
NM_007501
|
Neurod4
|
neurogenic differentiation 4
|
|
chr1_-_193399352
|
1.654
|
|
Dtl
|
denticleless homolog (Drosophila)
|
|
chr5_-_151468105
|
1.649
|
NM_201369
|
N4bp2l2
|
NEDD4 binding protein 2-like 2
|
|
chr1_-_59820157
|
1.625
|
|
B430105G09Rik
|
RIKEN cDNA B430105G09 gene
|
|
chr18_+_74893369
|
1.617
|
|
Myo5b
|
myosin VB
|
|
chr7_-_116867284
|
1.615
|
NM_001040700
|
BC051019
|
cDNA sequence BC051019
|
|
chr5_+_77739506
|
1.583
|
NM_153798
|
Polr2b
|
polymerase (RNA) II (DNA directed) polypeptide B
|
|
chr14_-_57190668
|
1.580
|
NM_001014996
|
Cenpj
|
centromere protein J
|
|
chr3_-_82707816
|
1.579
|
|
Lrat
|
lecithin-retinol acyltransferase (phosphatidylcholine-retinol-O-acyltransferase)
|
|
chr7_-_26022753
|
1.575
|
NM_001031667
|
Gsk3a
|
glycogen synthase kinase 3 alpha
|
|
chr7_+_95578782
|
1.573
|
NM_028238
|
Rab38
|
RAB38, member of RAS oncogene family
|
|
chr4_-_155571450
|
1.565
|
|
Agrn
|
agrin
|
|
chr7_+_129008551
|
1.562
|
NM_011325
|
Scnn1b
|
sodium channel, nonvoltage-gated 1 beta
|
|
chr11_-_120314753
|
1.562
|
NM_012065
|
Pde6g
|
phosphodiesterase 6G, cGMP-specific, rod, gamma
|
|
chr9_+_106724228
|
1.559
|
NM_001015507
|
Vprbp
|
Vpr (HIV-1) binding protein
|
|
chr7_-_138289796
|
1.552
|
NM_173409
|
4933402N03Rik
|
RIKEN cDNA 4933402N03 gene
|
|
chr13_-_67461473
|
1.545
|
NM_001038651
|
Zfp953
|
zinc finger protein 953
|
|
chr13_-_67370003
|
1.526
|
NM_001001152
|
Zfp458
|
zinc finger protein 458
|
|
chr14_+_49686169
|
1.525
|
NM_144535
|
Mudeng
|
MU-2/AP1M2 domain containing, death-inducing
|
|
chr12_+_37108036
|
1.507
|
NM_178629
|
Ispd
|
isoprenoid synthase domain containing
|
|
chr1_+_133724553
|
1.504
|
NM_173865
|
Slc41a1
|
solute carrier family 41, member 1
|
|
chr19_-_46747463
|
1.502
|
NM_007809
|
Cyp17a1
|
cytochrome P450, family 17, subfamily a, polypeptide 1
|
|
chr9_+_43915391
|
1.497
|
NM_001190319
|
C1qtnf5
|
C1q and tumor necrosis factor related protein 5
|
|
chr13_+_21570775
|
1.496
|
NM_001039115
|
Zkscan4
|
zinc finger with KRAB and SCAN domains 4
|
|
chr19_+_58803376
|
1.492
|
NM_018874
|
Pnliprp1
|
pancreatic lipase related protein 1
|
|
chr7_+_51357661
|
1.462
|
NM_010642
|
Klk1b21
|
kallikrein 1-related peptidase b21
|
|
chr2_+_154132962
|
1.449
|
NM_001025574
|
5430413K10Rik
|
RIKEN cDNA 5430413K10 gene
|
|
chr7_+_6124083
|
1.445
|
NM_001146024
NM_028316
|
Zfp444
|
zinc finger protein 444
|
|
chr4_+_106851234
|
1.444
|
NM_029565
|
Tmem59
|
transmembrane protein 59
|
|
chrX_-_101866123
|
1.431
|
|
Zdhhc15
|
zinc finger, DHHC domain containing 15
|
|
chr14_-_56503083
|
1.426
|
NM_019820
|
Cbln3
|
cerebellin 3 precursor protein
|
|
chr12_+_74040868
|
1.425
|
NM_011384
|
Six6
|
sine oculis-related homeobox 6 homolog (Drosophila)
|
|
chr18_-_32299223
|
1.425
|
NM_001042767
|
Proc
|
protein C
|
|
chr7_-_52003986
|
1.417
|
NM_178734
|
Zfp473
|
zinc finger protein 473
|
|
chr13_-_67162084
|
1.414
|
NM_001166218
|
Zfp712
|
zinc finger protein 712
|
|
chr2_+_164228640
|
1.395
|
NM_009036
|
Rbpjl
|
recombination signal binding protein for immunoglobulin kappa J region-like
|
|
chr7_+_4971977
|
1.395
|
NM_001033383
|
Zfp865
|
zinc finger protein 865
|
|
chr14_+_35487052
|
1.394
|
NM_001004436
|
Wapal
|
wings apart-like homolog (Drosophila)
|
|
chr6_-_122436294
|
1.390
|
NM_027664
|
Rimklb
|
ribosomal modification protein rimK-like family member B
|
|
chr2_-_156712810
|
1.390
|
NM_029983
|
Sla2
|
Src-like-adaptor 2
|
|
chr2_-_164571430
|
1.386
|
|
Wfdc3
|
WAP four-disulfide core domain 3
|
|
chr16_-_36367622
|
1.382
|
NM_001082546
|
BC100530
BC117090
|
cDNA sequence BC100530
cDNA sequence BC1179090
|
|
chr10_-_13588578
|
1.382
|
NM_025446
|
Aig1
|
androgen-induced 1
|
|
chr9_+_64659238
|
1.380
|
|
Dennd4a
|
DENN/MADD domain containing 4A
|
|
chr13_-_67407347
|
1.363
|
NM_001003666
|
Zfp457
|
zinc finger protein 457
|
|
chr2_+_154439056
|
1.351
|
NM_199304
|
Zfp341
|
zinc finger protein 341
|
|
chr15_-_75812322
|
1.347
|
|
2410075B13Rik
|
RIKEN cDNA 2410075B13 gene
|
|
chr7_-_135381486
|
1.330
|
NM_177001
|
9130023H24Rik
|
RIKEN cDNA 9130023H24 gene
|
|
chr8_+_77517536
|
1.319
|
NM_178017
|
Hmgxb4
|
HMG box domain containing 4
|
|
chr19_+_7457020
|
1.317
|
|
1700105P06Rik
|
RIKEN cDNA 1700105P06 gene
|
|
chr7_-_30638635
|
1.313
|
NM_173738
|
Zfp940
|
zinc finger protein 940
|
|
chr2_+_22923653
|
1.305
|
NM_001102436
NM_028793
|
Acbd5
|
acyl-Coenzyme A binding domain containing 5
|
|
chr12_-_113146249
|
1.301
|
|
Ppp1r13b
|
protein phosphatase 1, regulatory (inhibitor) subunit 13B
|
|
chr16_+_13903253
|
1.293
|
|
Mpv17l
|
Mpv17 transgene, kidney disease mutant-like
|
|
chr8_+_72566711
|
1.289
|
NM_027481
|
Sugp1
|
SURP and G patch domain containing 1
|
|
chrX_+_6645338
|
1.289
|
NM_001042542
|
Akap4
|
A kinase (PRKA) anchor protein 4
|
|
chrX_+_136219520
|
1.282
|
NM_001081499
|
Tbc1d8b
|
TBC1 domain family, member 8B
|
|
chr17_-_79336374
|
1.278
|
NM_001024806
|
Cebpz
|
CCAAT/enhancer binding protein zeta
|
|
chr3_-_53267127
|
1.268
|
NM_172501
|
Nhlrc3
|
NHL repeat containing 3
|
|
chr15_-_98638348
|
1.267
|
NM_001013741
|
Ddn
|
dendrin
|
|
chr1_+_132697478
|
1.262
|
NM_001170632
NM_144960
|
Fcamr
|
Fc receptor, IgA, IgM, high affinity
|
|
chr18_-_34738725
|
1.254
|
NM_080637
|
Nme5
|
non-metastatic cells 5, protein expressed in (nucleoside-diphosphate kinase)
|
|
chr2_-_34813680
|
1.243
|
|
Traf1
|
TNF receptor-associated factor 1
|
|
chr1_+_195128163
|
1.233
|
NM_008484
|
Lamb3
|
laminin, beta 3
|
|
chrX_-_35988248
|
1.232
|
|
C1galt1c1
|
C1GALT1-specific chaperone 1
|
|
chr2_-_34813633
|
1.227
|
|
Traf1
|
TNF receptor-associated factor 1
|
|
chr11_+_14499242
|
1.226
|
NM_001164166
|
Pom121l12
|
POM121 membrane glycoprotein-like 12
|
|
chr8_+_72566937
|
1.225
|
|
Sugp1
|
SURP and G patch domain containing 1
|
|
chrX_-_159987479
|
1.221
|
NM_008177
|
Grpr
|
gastrin releasing peptide receptor
|
|
chr11_-_3763899
|
1.217
|
NM_152818
|
Osbp2
|
oxysterol binding protein 2
|
|
chr5_+_115777174
|
1.215
|
NM_025573
|
Srsf9
|
serine/arginine-rich splicing factor 9
|
|
chrX_+_7156801
|
1.211
|
NM_054039
|
Foxp3
|
forkhead box P3
|
|
chr8_+_72346327
|
1.205
|
NM_020028
|
Lpar2
|
lysophosphatidic acid receptor 2
|
|
chr11_+_68970573
|
1.201
|
NM_009659
|
Alox12b
|
arachidonate 12-lipoxygenase, 12R type
|
|
chr17_-_47147501
|
1.196
|
|
Ubr2
C79242
|
ubiquitin protein ligase E3 component n-recognin 2
expressed sequence C79242
|
|
chr15_-_54903657
|
1.189
|
NM_001081288
|
Taf2
|
TAF2 RNA polymerase II, TATA box binding protein (TBP)-associated factor
|
|
chr2_-_92210955
|
1.187
|
NM_001166633
NM_172670
|
Gyltl1b
|
glycosyltransferase-like 1B
|
|
chr9_+_107830799
|
1.185
|
NM_026338
|
4921517D21Rik
|
RIKEN cDNA 4921517D21 gene
|
|
chr8_+_19784703
|
1.169
|
|
2610005L07Rik
A430108E01Rik
|
cadherin 11 pseudogene
RIKEN cDNA A430108E01 gene
|
|
chr9_+_107790512
|
1.165
|
|
Mon1a
|
MON1 homolog A (yeast)
|
|
chr10_+_21097094
|
1.162
|
NM_178713
|
Aldh8a1
|
aldehyde dehydrogenase 8 family, member A1
|
|
chr14_-_56503058
|
1.161
|
|
Cbln3
|
cerebellin 3 precursor protein
|
|
chr6_-_148894953
|
1.160
|
NM_019643
|
Fam60a
|
family with sequence similarity 60, member A
|
|
chr19_+_32463705
|
1.160
|
|
2700046G09Rik
|
RIKEN cDNA 2700046G09 gene
|
|
chr7_-_6649139
|
1.158
|
NM_011769
|
Zim1
|
zinc finger, imprinted 1
|
|
chr5_+_73039082
|
1.155
|
|
Nipal1
|
NIPA-like domain containing 1
|
|
chr4_+_40419212
|
1.154
|
NM_177175
|
Tmem215
|
transmembrane protein 215
|
|
chr16_-_55838747
|
1.147
|
NM_001159395
NM_030612
|
Nfkbiz
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, zeta
|
|
chr7_+_135381536
|
1.139
|
|
Armc5
|
armadillo repeat containing 5
|
|
chr18_-_34784206
|
1.139
|
|
Brd8
|
bromodomain containing 8
|
|
chr12_+_8797502
|
1.136
|
|
|
|
|
chr2_-_165060352
|
1.122
|
|
Cdh22
|
cadherin 22
|
|
chr14_+_31699648
|
1.121
|
NM_001159299
NM_018746
|
Itih4
|
inter alpha-trypsin inhibitor, heavy chain 4
|
|
chr15_-_34608458
|
1.119
|
NM_145469
|
Nipal2
|
NIPA-like domain containing 2
|
|
chr18_+_31793991
|
1.116
|
NM_172965
|
Sap130
|
Sin3A associated protein
|
|
chr6_+_146798631
|
1.115
|
NM_027059
|
1700023A16Rik
|
RIKEN cDNA 1700023A16 gene
|
|
chr6_-_85854877
|
1.110
|
NM_029331
|
1700019G17Rik
|
RIKEN cDNA 1700019G17 gene
|
|
chr6_-_91713072
|
1.105
|
|
Grip2
|
glutamate receptor interacting protein 2
|
|
chr9_+_27204405
|
1.102
|
NM_029299
|
Spata19
|
spermatogenesis associated 19
|
|
chr6_+_21899614
|
1.099
|
NM_023626
|
Ing3
|
inhibitor of growth family, member 3
|
|
chr10_+_120834091
|
1.087
|
|
|
|
|
chr16_+_96502595
|
1.086
|
NM_033149
|
B3galt5
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 5
|
|
chr1_-_193399412
|
1.084
|
NM_029766
|
Dtl
|
denticleless homolog (Drosophila)
|
|
chr8_+_19784580
|
1.083
|
|
A430108E01Rik
|
RIKEN cDNA A430108E01 gene
|
|
chr9_-_50554613
|
1.077
|
NM_025865
|
2310030G06Rik
|
RIKEN cDNA 2310030G06 gene
|
|
chr5_+_3543815
|
1.075
|
NM_001042501
|
Fam133b
|
family with sequence similarity 133, member B
|
|
chr11_+_78317000
|
1.072
|
NM_008759
|
Sebox
|
SEBOX homeobox
|
|
chr11_+_113545719
|
1.070
|
NM_177777
|
D11Wsu47e
|
DNA segment, Chr 11, Wayne State University 47, expressed
|
|
chrX_+_96331434
|
1.070
|
NM_010110
|
Efnb1
|
ephrin B1
|
|
chr6_+_78375973
|
1.065
|
NM_009042
|
Reg1
|
regenerating islet-derived 1
|
|
chr9_-_50369372
|
1.065
|
NM_025687
|
Tex12
|
testis expressed gene 12
|
|
chr7_+_86418127
|
1.063
|
NM_018811
|
Abhd2
|
abhydrolase domain containing 2
|
|
chr1_-_129574328
|
1.060
|
|
Tmem163
|
transmembrane protein 163
|
|
chr2_-_60511707
|
1.060
|
NM_021359
|
Itgb6
|
integrin beta 6
|
|
chr6_-_23600205
|
1.058
|
NM_198251
|
Rnf133
|
ring finger protein 133
|
|
chr4_-_22415444
|
1.057
|
|
Pou3f2
|
POU domain, class 3, transcription factor 2
|
|
chr15_-_98638230
|
1.053
|
|
Ddn
|
dendrin
|