|
chr12_-_73509668
|
13.015
|
|
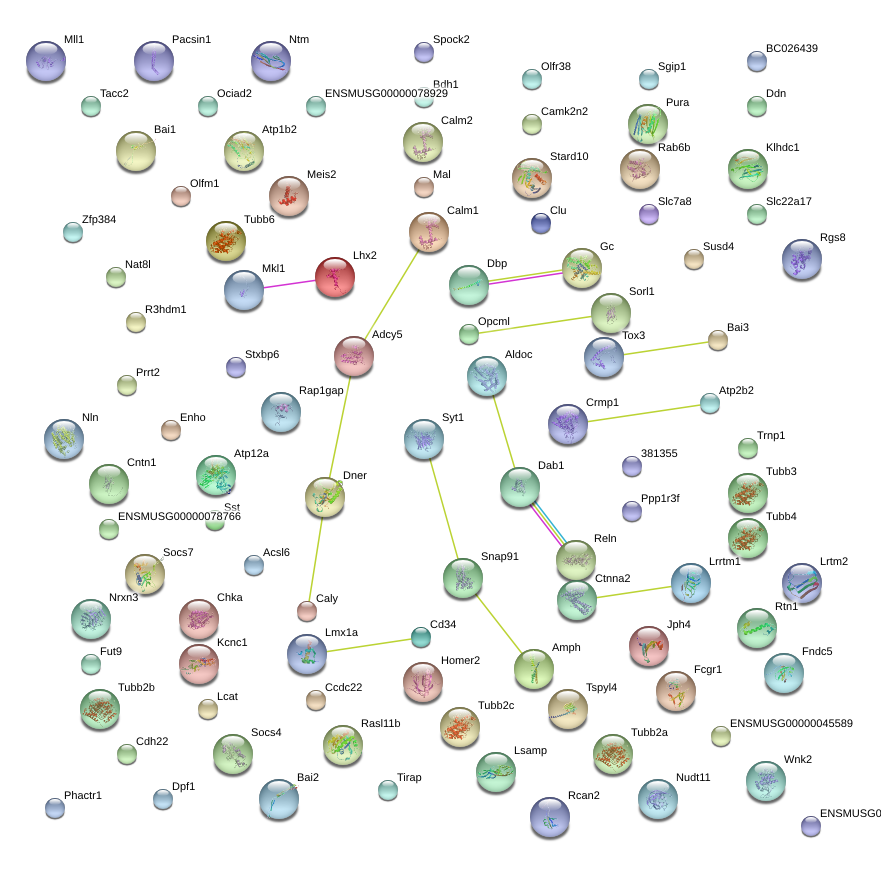
Rtn1
|
reticulon 1
|
|
chr12_-_73509537
|
11.675
|
|
Rtn1
|
reticulon 1
|
|
chr14_+_66587319
|
10.656
|
NM_013492
|
Clu
|
clusterin
|
|
chr15_+_91881593
|
10.069
|
NM_001159648
|
Cntn1
|
contactin 1
|
|
chr4_+_129662284
|
9.334
|
NM_173071
|
Bai2
|
brain-specific angiogenesis inhibitor 2
|
|
chr4_+_104040160
|
8.583
|
NM_010014
NM_177259
|
Dab1
|
disabled homolog 1 (Drosophila)
|
|
chr9_-_29219382
|
8.122
|
|
Ntm
|
neurotrimin
|
|
chr9_-_86775282
|
8.032
|
|
Snap91
|
synaptosomal-associated protein 91
|
|
chr10_-_108447894
|
7.911
|
|
Syt1
|
synaptotagmin I
|
|
chr9_-_86775154
|
7.819
|
NM_013669
|
Snap91
|
synaptosomal-associated protein 91
|
|
chr11_+_98629709
|
7.657
|
|
|
|
|
chr2_+_28048779
|
7.515
|
|
Olfm1
|
olfactomedin 1
|
|
chr10_-_108448003
|
6.976
|
|
Syt1
|
synaptotagmin I
|
|
chr4_+_129662110
|
6.972
|
|
Bai2
|
brain-specific angiogenesis inhibitor 2
|
|
chr6_-_119280616
|
6.865
|
NM_172492
|
Lrtm2
|
leucine-rich repeats and transmembrane domains 2
|
|
chr14_-_55734213
|
6.654
|
NM_001003829
|
Jph4
|
junctophilin 4
|
|
chr4_-_41587115
|
6.640
|
|
Enho
|
energy homeostasis associated
|
|
chr6_-_113841295
|
6.534
|
|
Atp2b2
|
ATPase, Ca++ transporting, plasma membrane 2
|
|
chr7_+_53651811
|
6.517
|
NM_008421
NM_001112739
|
Kcnc1
|
potassium voltage gated channel, Shaw-related subfamily, member 1
|
|
chr9_-_44688946
|
6.319
|
|
Mll1
|
myeloid/lymphoid or mixed-lineage leukemia 1
|
|
chr5_+_74591336
|
6.225
|
NM_026878
|
Rasl11b
|
RAS-like, family 11, member B
|
|
chr7_-_88851823
|
6.126
|
|
Homer2
|
homer homolog 2 (Drosophila)
|
|
chr9_+_27598853
|
6.081
|
NM_177906
|
Opcml
|
opioid binding protein/cell adhesion molecule-like
|
|
chr1_-_84692352
|
6.057
|
|
Dner
|
delta/notch-like EGF-related receptor
|
|
chr13_-_49243351
|
6.052
|
NM_029361
|
Wnk2
|
WNK lysine deficient protein kinase 2
|
|
chr9_+_103014374
|
5.899
|
NM_173781
|
Rab6b
|
RAB6B, member RAS oncogene family
|
|
chr12_-_73509925
|
5.807
|
NM_153457
|
Rtn1
|
reticulon 1
|
|
chr6_-_113841356
|
5.772
|
NM_001036684
|
Atp2b2
|
ATPase, Ca++ transporting, plasma membrane 2
|
|
chr15_-_98637560
|
5.682
|
|
Ddn
|
dendrin
|
|
chr5_-_21850490
|
5.680
|
NM_011261
|
Reln
|
reelin
|
|
chr2_+_38206759
|
5.659
|
NM_010710
|
Lhx2
|
LIM homeobox protein 2
|
|
chr7_+_108469700
|
5.614
|
|
Stard10
|
START domain containing 10
|
|
chr11_-_69419330
|
5.534
|
|
Atp1b2
|
ATPase, Na+/K+ transporting, beta 2 polypeptide
|
|
chr2_-_115890247
|
5.493
|
NM_001159569
NM_001159570
|
Meis2
|
Meis homeobox 2
|
|
chr10_+_59569206
|
5.476
|
|
Spock2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan 2
|
|
chr13_+_19040215
|
5.439
|
NM_175007
|
Amph
|
amphiphysin
|
|
chr5_+_34339258
|
5.433
|
|
Nat8l
|
N-acetyltransferase 8-like
|
|
chr6_+_77193078
|
5.402
|
|
Lrrtm1
|
leucine rich repeat transmembrane neuronal 1
|
|
chr14_-_55531726
|
5.374
|
NM_021551
|
Slc22a17
|
solute carrier family 22 (organic cation transporter), member 17
|
|
chr1_+_155500166
|
5.357
|
NM_026380
|
Rgs8
|
regulator of G-protein signaling 8
|
|
chr17_+_27792487
|
5.337
|
|
Pacsin1
|
protein kinase C and casein kinase substrate in neurons 1
|
|
chr12_+_101447480
|
5.302
|
|
Calm1
|
calmodulin 1
|
|
chrX_+_5624624
|
5.261
|
NM_021431
|
Nudt11
|
nudix (nucleoside diphosphate linked moiety X)-type motif 11
|
|
chr16_-_20621287
|
5.139
|
NM_028420
|
Camk2n2
|
calcium/calmodulin-dependent protein kinase II inhibitor 2
|
|
chr16_+_35154935
|
5.126
|
|
Adcy5
|
adenylate cyclase 5
|
|
chr6_-_77929685
|
5.099
|
|
Ctnna2
|
catenin (cadherin associated protein), alpha 2
|
|
chr2_+_38194775
|
5.098
|
|
Lhx2
|
LIM homeobox protein 2
|
|
chr18_+_36446680
|
5.025
|
|
Pura
|
purine rich element binding protein A
|
|
chr1_+_184695026
|
4.969
|
NM_144796
|
Susd4
|
sushi domain containing 4
|
|
chr7_-_147268150
|
4.916
|
NM_001190386
|
Caly
|
calcyon neuron-specific vesicular protein
|
|
chr7_-_134164627
|
4.914
|
NM_001102563
|
Prrt2
|
proline-rich transmembrane protein 2
|
|
chr12_+_91050912
|
4.877
|
|
Nrxn3
|
neurexin III
|
|
chr9_-_41932371
|
4.821
|
NM_011436
|
Sorl1
|
sortilin-related receptor, LDLR class A repeats-containing
|
|
chr17_-_57226987
|
4.765
|
NM_009451
|
Tubb4
|
tubulin, beta 4
|
|
chr4_-_25727109
|
4.750
|
NM_010243
|
Fut9
|
fucosyltransferase 9
|
|
chr12_-_46175433
|
4.739
|
NM_144552
|
Stxbp6
|
syntaxin binding protein 6 (amisyn)
|
|
chr9_-_41932195
|
4.699
|
|
Sorl1
|
sortilin-related receptor, LDLR class A repeats-containing
|
|
chr5_+_34339351
|
4.680
|
|
Nat8l
|
N-acetyltransferase 8-like
|
|
chr11_+_54117703
|
4.600
|
NM_001033597
NM_144823
|
Acsl6
|
acyl-CoA synthetase long-chain family member 6
|
|
chr14_-_55400722
|
4.599
|
NM_016972
|
Slc7a8
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 8
|
|
chr1_+_129999868
|
4.542
|
NM_181750
|
R3hdm1
|
R3H domain 1 (binds single-stranded nucleic acids)
|
|
chr8_-_92872139
|
4.540
|
NM_172913
|
Tox3
|
TOX high mobility group box family member 3
|
|
chr19_+_3851772
|
4.391
|
NM_001025566
NM_013490
|
Chka
|
choline kinase alpha
|
|
chr13_+_43218207
|
4.333
|
NM_001005748
|
Phactr1
|
phosphatase and actin regulator 1
|
|
chr11_+_78136681
|
4.324
|
|
Aldoc
|
aldolase C, fructose-bisphosphate
|
|
chr2_+_28048592
|
4.306
|
NM_001038613
NM_001038614
|
Olfm1
|
olfactomedin 1
|
|
chr11_+_54128404
|
4.258
|
NM_001033599
|
Acsl6
|
acyl-CoA synthetase long-chain family member 6
|
|
chr7_+_137789668
|
4.253
|
|
Tacc2
|
transforming, acidic coiled-coil containing protein 2
|
|
chr2_-_127482430
|
4.252
|
NM_001171187
NM_010762
|
Mal
|
myelin and lymphocyte protein, T-cell differentiation protein
|
|
chr4_-_57002965
|
4.230
|
|
6430704M03Rik
|
RIKEN cDNA 6430704M03 gene
|
|
chr5_-_73729789
|
4.204
|
NM_026950
|
Ociad2
|
OCIA domain containing 2
|
|
chr16_-_23890909
|
4.184
|
NM_009215
|
Sst
|
somatostatin
|
|
chr2_-_165060352
|
4.175
|
|
Cdh22
|
cadherin 22
|
|
chr17_+_43938789
|
4.142
|
NM_207649
|
Rcan2
|
regulator of calcineurin 2
|
|
chrX_+_5624820
|
4.140
|
|
Nudt11
|
nudix (nucleoside diphosphate linked moiety X)-type motif 11
|
|
chr8_-_108467294
|
4.112
|
NM_008490
|
Lcat
|
lecithin cholesterol acyltransferase
|
|
chr6_+_124959207
|
4.093
|
|
Zfp384
|
zinc finger protein 384
|
|
chr7_+_30088984
|
4.084
|
NM_013874
|
Dpf1
|
D4, zinc and double PHD fingers family 1
|
|
chr4_+_128820458
|
4.023
|
|
Fndc5
|
fibronectin type III domain containing 5
|
|
chr4_+_102432879
|
4.018
|
|
Sgip1
|
SH3-domain GRB2-like (endophilin) interacting protein 1
|
|
chr16_+_41533317
|
4.018
|
NM_175548
|
Lsamp
|
limbic system-associated membrane protein
|
|
chr11_+_97258202
|
4.007
|
|
Socs7
|
suppressor of cytokine signaling 7
|
|
chr12_+_70342818
|
3.908
|
NM_178253
|
Klhdc1
|
kelch domain containing 1
|
|
chr11_+_97223739
|
3.905
|
|
Socs7
|
suppressor of cytokine signaling 7
|
|
chr4_-_133054355
|
3.875
|
NM_001081156
|
Trnp1
|
TMF1-regulated nuclear protein 1
|
|
chr7_+_52960585
|
3.836
|
NM_016974
|
Dbp
|
D site albumin promoter binding protein
|
|
chrX_-_7151340
|
3.828
|
|
Ppp1r3f
|
protein phosphatase 1, regulatory (inhibitor) subunit 3F
|
|
chr16_+_31428887
|
3.823
|
|
Bdh1
|
3-hydroxybutyrate dehydrogenase, type 1
|
|
chr4_+_137220675
|
3.793
|
|
Rap1gap
|
Rap1 GTPase-activating protein
|
|
chr10_+_34017221
|
3.769
|
NM_030203
|
Tspyl4
|
TSPY-like 4
|
|
chr8_-_108467275
|
3.725
|
|
Lcat
|
lecithin cholesterol acyltransferase
|
|
chr11_+_32183671
|
3.723
|
NM_001083955
NM_008218
|
Hba-a2
Hba-a1
|
hemoglobin alpha, adult chain 2
hemoglobin alpha, adult chain 1
|
|
chr6_+_4853319
|
3.718
|
NM_181595
|
Ppp1r9a
|
protein phosphatase 1, regulatory (inhibitor) subunit 9A
|
|
chr2_+_164312023
|
3.713
|
|
Dbndd2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2
|
|
chr13_+_94074449
|
3.707
|
NM_011982
NM_147176
|
Homer1
|
homer homolog 1 (Drosophila)
|
|
chr9_-_83699706
|
3.681
|
NM_001145974
NM_148941
|
Elovl4
|
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 4
|
|
chr17_+_25707771
|
3.674
|
|
2810468N07Rik
|
RIKEN cDNA 2810468N07 gene
|
|
chr7_-_88851798
|
3.670
|
NM_001164086
NM_011983
|
Homer2
|
homer homolog 2 (Drosophila)
|
|
chr1_+_171585621
|
3.656
|
NM_009063
|
Rgs5
|
regulator of G-protein signaling 5
|
|
chr15_-_101834746
|
3.652
|
NM_031170
|
Krt8
|
keratin 8
|
|
chr9_+_103014279
|
3.641
|
|
Rab6b
|
RAB6B, member RAS oncogene family
|
|
chr1_+_75233276
|
3.627
|
NM_001159885
|
Dnajb2
|
DnaJ (Hsp40) homolog, subfamily B, member 2
|
|
chr6_-_77929543
|
3.623
|
|
Ctnna2
|
catenin (cadherin associated protein), alpha 2
|
|
chr1_-_84692750
|
3.614
|
NM_152915
|
Dner
|
delta/notch-like EGF-related receptor
|
|
chr1_-_134638361
|
3.609
|
NM_001160317
NM_182716
|
Nfasc
|
neurofascin
|
|
chr13_+_42775991
|
3.602
|
NM_198419
|
Phactr1
|
phosphatase and actin regulator 1
|
|
chr4_-_150503464
|
3.602
|
|
Camta1
|
calmodulin binding transcription activator 1
|
|
chr2_-_148558110
|
3.588
|
NM_019632
|
Napb
|
N-ethylmaleimide sensitive fusion protein attachment protein beta
|
|
chr11_+_68901989
|
3.584
|
|
Vamp2
|
vesicle-associated membrane protein 2
|
|
chr9_+_44947045
|
3.577
|
NM_001013390
|
Scn4b
|
sodium channel, type IV, beta
|
|
chr1_+_109258889
|
3.574
|
NM_025867
|
Serpinb11
|
serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 11
|
|
chr3_-_89577134
|
3.567
|
|
4632404H12Rik
|
RIKEN cDNA 4632404H12 gene
|
|
chr7_+_134094045
|
3.565
|
NM_144926
|
Sez6l2
|
seizure related 6 homolog like 2
|
|
chr5_+_37633353
|
3.536
|
|
Crmp1
|
collapsin response mediator protein 1
|
|
chr17_-_57199207
|
3.534
|
NM_025877
|
Slc25a23
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 23
|
|
chr4_-_122638401
|
3.526
|
NM_029662
|
Mfsd2a
|
major facilitator superfamily domain containing 2A
|
|
chr19_+_5689759
|
3.521
|
|
Map3k11
|
mitogen-activated protein kinase kinase kinase 11
|
|
chr2_+_155343804
|
3.515
|
|
Acss2
|
acyl-CoA synthetase short-chain family member 2
|
|
chr1_-_75274936
|
3.506
|
NM_009049
|
Resp18
|
regulated endocrine-specific protein 18
|
|
chr1_-_75274890
|
3.498
|
|
Resp18
|
regulated endocrine-specific protein 18
|
|
chr8_+_32200076
|
3.493
|
NM_025869
|
Dusp26
|
dual specificity phosphatase 26 (putative)
|
|
chr2_+_29658139
|
3.485
|
NM_011989
|
Slc27a4
|
solute carrier family 27 (fatty acid transporter), member 4
|
|
chr4_+_102432943
|
3.465
|
NM_144906
|
Sgip1
|
SH3-domain GRB2-like (endophilin) interacting protein 1
|
|
chr10_+_122115755
|
3.450
|
NM_001110218
NM_176919
|
Ppm1h
|
protein phosphatase 1H (PP2C domain containing)
|
|
chr15_-_83936038
|
3.440
|
|
Sult4a1
|
sulfotransferase family 4A, member 1
|
|
chr5_-_31395853
|
3.438
|
NM_011773
|
Slc30a3
|
solute carrier family 30 (zinc transporter), member 3
|
|
chr2_-_180891448
|
3.426
|
|
Eef1a2
|
eukaryotic translation elongation factor 1 alpha 2
|
|
chr1_+_50984361
|
3.390
|
NM_019790
|
Tmeff2
|
transmembrane protein with EGF-like and two follistatin-like domains 2
|
|
chr7_-_135073617
|
3.371
|
NM_133351
|
Prss8
|
protease, serine, 8 (prostasin)
|
|
chr11_+_102622885
|
3.368
|
|
Adam11
|
a disintegrin and metallopeptidase domain 11
|
|
chr18_-_31476527
|
3.362
|
|
Rit2
|
Ras-like without CAAX 2
|
|
chr15_-_98638230
|
3.359
|
|
Ddn
|
dendrin
|
|
chr8_+_65430028
|
3.349
|
NM_023689
|
Spock3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan 3
|
|
chr5_-_121161653
|
3.336
|
|
Dtx1
|
deltex 1 homolog (Drosophila)
|
|
chr9_+_108998083
|
3.335
|
|
Plxnb1
|
plexin B1
|
|
chr16_+_7069927
|
3.332
|
NM_183188
|
Rbfox1
|
RNA binding protein, fox-1 homolog (C. elegans) 1
|
|
chr19_+_47089168
|
3.323
|
NM_146100
|
Ina
|
internexin neuronal intermediate filament protein, alpha
|
|
chr4_-_124539414
|
3.319
|
NM_001007573
|
Maneal
|
mannosidase, endo-alpha-like
|
|
chr7_+_48154647
|
3.318
|
NM_021387
|
Vstm2b
|
V-set and transmembrane domain containing 2B
|
|
chr19_+_48280514
|
3.309
|
NM_025696
|
Sorcs3
|
sortilin-related VPS10 domain containing receptor 3
|
|
chr4_-_68615283
|
3.281
|
|
Dbc1
|
deleted in bladder cancer 1 (human)
|
|
chr5_+_34339551
|
3.277
|
|
Nat8l
|
N-acetyltransferase 8-like
|
|
chr6_+_53989925
|
3.272
|
NM_001163640
|
Chn2
|
chimerin (chimaerin) 2
|
|
chr7_-_104886507
|
3.270
|
|
Aqp11
|
aquaporin 11
|
|
chr15_-_101834744
|
3.256
|
|
Krt8
|
keratin 8
|
|
chr18_+_64425037
|
3.241
|
|
St8sia3
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3
|
|
chr12_+_81619976
|
3.201
|
NM_001081421
|
Galntl1
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 1
|
|
chr7_+_52960702
|
3.183
|
|
Dbp
|
D site albumin promoter binding protein
|
|
chr2_-_168567422
|
3.182
|
|
Atp9a
|
ATPase, class II, type 9A
|
|
chr7_-_25772525
|
3.181
|
|
Atp1a3
|
ATPase, Na+/K+ transporting, alpha 3 polypeptide
|
|
chr14_-_76956445
|
3.167
|
NM_001160326
|
Serp2
|
stress-associated endoplasmic reticulum protein family member 2
|
|
chr9_+_103014445
|
3.166
|
|
Rab6b
|
RAB6B, member RAS oncogene family
|
|
chr4_-_68615382
|
3.138
|
NM_019967
|
Dbc1
|
deleted in bladder cancer 1 (human)
|
|
chr9_-_35365985
|
3.136
|
NM_013932
|
Ddx25
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 25
|
|
chr1_+_66515003
|
3.119
|
NM_175510
|
Unc80
|
unc-80 homolog (C. elegans)
|
|
chr7_-_135073600
|
3.113
|
|
Prss8
|
protease, serine, 8 (prostasin)
|
|
chr2_-_165060188
|
3.112
|
NM_174988
|
Cdh22
|
cadherin 22
|
|
chr7_-_30066867
|
3.104
|
|
Spint2
|
serine protease inhibitor, Kunitz type 2
|
|
chr2_+_149656518
|
3.084
|
NM_001085521
|
Tmem90b
|
transmembrane protein 90B
|
|
chr11_-_64976408
|
3.061
|
NM_001099288
NM_175003
|
Arhgap44
|
Rho GTPase activating protein 44
|
|
chr5_-_118694869
|
3.049
|
NM_172998
NM_001109902
|
Rnft2
|
ring finger protein, transmembrane 2
|
|
chr2_+_102498839
|
3.047
|
NM_001077514
|
Slc1a2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2
|
|
chr7_-_31319977
|
3.021
|
NM_178252
|
Arhgap33
|
Rho GTPase activating protein 33
|
|
chr1_+_74379182
|
3.012
|
NM_019999
|
Pnkd
|
paroxysmal nonkinesiogenic dyskinesia
|
|
chr7_+_134716853
|
3.011
|
NM_026888
|
Phkg2
|
phosphorylase kinase, gamma 2 (testis)
|
|
chr4_+_122860705
|
3.009
|
NM_174998
|
Hpcal4
|
hippocalcin-like 4
|
|
chr12_+_3365295
|
3.001
|
|
Kif3c
|
kinesin family member 3C
|
|
chr18_-_31476721
|
3.001
|
NM_009065
|
Rit2
|
Ras-like without CAAX 2
|
|
chr1_+_194014650
|
3.000
|
|
Kcnh1
|
potassium voltage-gated channel, subfamily H (eag-related), member 1
|
|
chr5_-_123582394
|
2.998
|
|
Rhof
|
ras homolog gene family, member f
|
|
chr11_-_116442760
|
2.996
|
NM_173755
|
Ube2o
|
ubiquitin-conjugating enzyme E2O
|
|
chr7_-_31758185
|
2.994
|
|
Lsr
|
lipolysis stimulated lipoprotein receptor
|
|
chr1_+_34858612
|
2.992
|
|
Arhgef4
|
Rho guanine nucleotide exchange factor (GEF) 4
|
|
chr7_-_26460133
|
2.992
|
|
Tmem91
|
transmembrane protein 91
|
|
chr14_-_46277816
|
2.992
|
NM_001039106
NM_001042719
NM_176845
|
Ddhd1
|
DDHD domain containing 1
|
|
chr11_-_98190774
|
2.990
|
NM_010895
|
Neurod2
|
neurogenic differentiation 2
|
|
chr5_-_53604673
|
2.986
|
NM_172710
|
Sel1l3
|
sel-1 suppressor of lin-12-like 3 (C. elegans)
|
|
chr3_-_108218411
|
2.982
|
NM_001004177
NM_017392
|
Celsr2
|
cadherin, EGF LAG seven-pass G-type receptor 2 (flamingo homolog, Drosophila)
|
|
chr1_-_166388431
|
2.970
|
NM_009721
|
Atp1b1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide
|
|
chr17_-_46865894
|
2.970
|
|
Gnmt
|
glycine N-methyltransferase
|
|
chr11_-_70501586
|
2.968
|
NM_001190376
NM_001190378
NM_001190379
NM_178116
|
Camta2
|
calmodulin binding transcription activator 2
|
|
chr14_-_13655592
|
2.958
|
NM_001042617
NM_012061
|
Cadps
|
Ca2+-dependent secretion activator
|
|
chr4_-_25726753
|
2.945
|
|
Fut9
|
fucosyltransferase 9
|
|
chr16_+_96689373
|
2.943
|
|
Pcp4
|
Purkinje cell protein 4
|
|
chrX_-_149471740
|
2.942
|
NM_023788
|
Mageh1
|
melanoma antigen, family H, 1
|
|
chr5_-_44485951
|
2.938
|
NM_001163578
NM_001163581
NM_001163582
NM_001163583
NM_001163584
NM_001163585
|
Prom1
|
prominin 1
|
|
chr2_+_25251393
|
2.938
|
NM_009849
|
Entpd2
|
ectonucleoside triphosphate diphosphohydrolase 2
|
|
chr11_-_94902584
|
2.932
|
NM_133667
|
Pdk2
|
pyruvate dehydrogenase kinase, isoenzyme 2
|
|
chr9_+_103014463
|
2.930
|
|
Rab6b
|
RAB6B, member RAS oncogene family
|
|
chr18_-_39078304
|
2.928
|
NM_010197
|
Fgf1
|
fibroblast growth factor 1
|
|
chr7_-_135073577
|
2.927
|
|
Prss8
|
protease, serine, 8 (prostasin)
|
|
chr18_+_56591785
|
2.923
|
NM_026240
|
Gramd3
|
GRAM domain containing 3
|
|
chrX_-_103414967
|
2.913
|
|
Taf9b
|
TAF9B RNA polymerase II, TATA box binding protein (TBP)-associated factor
|
|
chr7_-_107806887
|
2.902
|
NM_001163183
NM_001163184
NM_001163185
|
Plekhb1
|
pleckstrin homology domain containing, family B (evectins) member 1
|
|
chr1_-_193220140
|
2.893
|
|
|
|
|
chr11_-_4847968
|
2.881
|
NM_010904
|
Nefh
|
neurofilament, heavy polypeptide
|
|
chr14_-_13655558
|
2.870
|
|
Cadps
|
Ca2+-dependent secretion activator
|
|
chr11_-_72303447
|
2.870
|
|
Spns2
|
spinster homolog 2 (Drosophila)
|
|
chr12_-_112910475
|
2.864
|
NM_021273
|
Ckb
|
creatine kinase, brain
|
|
chr11_+_87394543
|
2.864
|
|
Sept4
|
septin 4
|