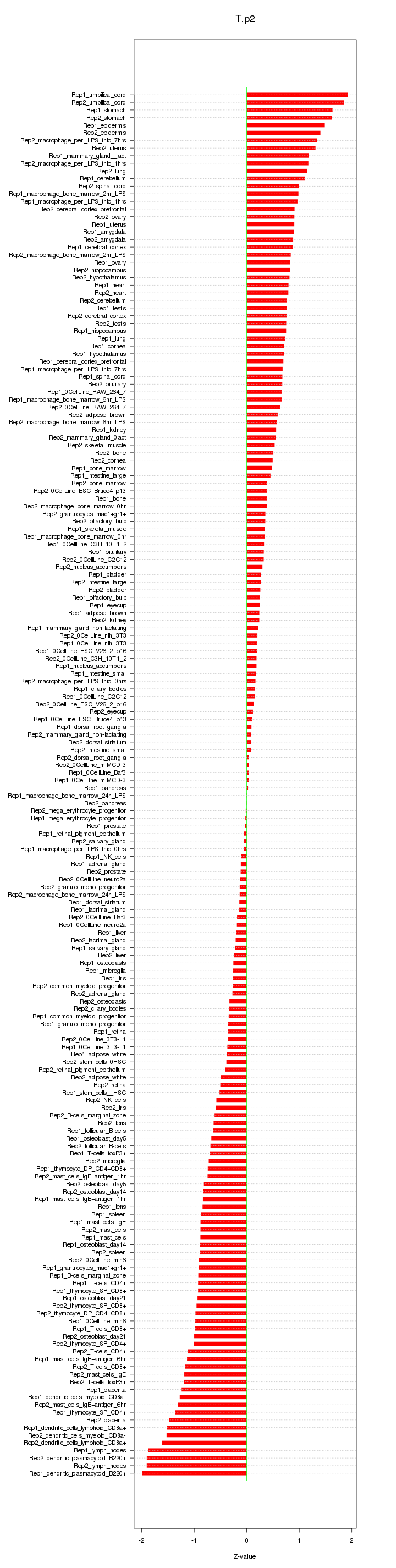
|
chr19_+_20676453
|
16.693
|
NM_013467
|
Aldh1a1
|
aldehyde dehydrogenase family 1, subfamily A1
|
|
chr15_+_12082671
|
12.584
|
|
Zfr
|
zinc finger RNA binding protein
|
|
chr7_-_107805466
|
12.225
|
NM_001163186
NM_001163187
|
Plekhb1
|
pleckstrin homology domain containing, family B (evectins) member 1
|
|
chr10_-_61738537
|
11.347
|
|
Hk1
|
hexokinase 1
|
|
chr8_+_46970860
|
9.873
|
|
Pdlim3
|
PDZ and LIM domain 3
|
|
chr8_+_46970814
|
9.862
|
NM_016798
|
Pdlim3
|
PDZ and LIM domain 3
|
|
chr15_-_101681183
|
9.372
|
NM_008473
|
Krt1
|
keratin 1
|
|
chr15_-_81191176
|
9.252
|
NM_011399
|
Slc25a17
|
solute carrier family 25 (mitochondrial carrier, peroxisomal membrane protein), member 17
|
|
chr15_-_12522310
|
9.018
|
NM_001081064
|
Pdzd2
|
PDZ domain containing 2
|
|
chr19_+_23240506
|
6.947
|
|
Klf9
|
Kruppel-like factor 9
|
|
chr12_+_110783593
|
6.694
|
|
Meg3
|
maternally expressed 3
|
|
chr11_-_55208276
|
6.677
|
|
|
|
|
chrX_-_147091911
|
6.634
|
NM_001002272
NM_019548
|
Tro
|
trophinin
|
|
chr5_-_143668369
|
6.597
|
|
Actb
|
actin, beta
|
|
chr7_-_107806887
|
6.433
|
NM_001163183
NM_001163184
NM_001163185
|
Plekhb1
|
pleckstrin homology domain containing, family B (evectins) member 1
|
|
chr8_-_126957835
|
6.401
|
NM_171824
|
Pgbd5
|
piggyBac transposable element derived 5
|
|
chr12_+_110806715
|
5.950
|
|
Meg3
|
maternally expressed 3
|
|
chr3_-_107820766
|
5.944
|
NM_010358
|
Gstm1
|
glutathione S-transferase, mu 1
|
|
chr2_-_25079684
|
5.609
|
|
Tubb2c
|
tubulin, beta 2C
|
|
chr7_-_4498168
|
5.254
|
NM_016908
|
Syt5
|
synaptotagmin V
|
|
chr6_-_24905939
|
5.167
|
NM_177013
|
Tmem229a
|
transmembrane protein 229A
|
|
chr4_-_82151657
|
5.124
|
|
Nfib
|
nuclear factor I/B
|
|
chr7_-_25772525
|
5.074
|
|
Atp1a3
|
ATPase, Na+/K+ transporting, alpha 3 polypeptide
|
|
chr12_+_110783786
|
4.871
|
|
Meg3
|
maternally expressed 3
|
|
chr3_+_67696141
|
4.777
|
NM_001113419
NM_177585
|
Iqcj-schip1
Iqcj
|
IQ motif containing J-schwannomin interacting protein 1 read-through transcript
IQ motif containing J
|
|
chr7_-_6683113
|
4.768
|
NM_008817
|
Peg3
|
paternally expressed 3
|
|
chr5_+_91201521
|
4.748
|
|
Pf4
|
platelet factor 4
|
|
chr10_-_24919916
|
4.583
|
|
Akap7
|
A kinase (PRKA) anchor protein 7
|
|
chr11_-_100599413
|
4.582
|
NM_024456
|
Rab5c
|
RAB5C, member RAS oncogene family
|
|
chr6_-_94650118
|
4.470
|
NM_008377
|
Lrig1
|
leucine-rich repeats and immunoglobulin-like domains 1
|
|
chr7_-_82453166
|
4.172
|
|
Sv2b
|
synaptic vesicle glycoprotein 2 b
|
|
chrX_+_133200982
|
4.097
|
|
Tceal3
|
transcription elongation factor A (SII)-like 3
|
|
chr4_-_75779379
|
3.940
|
NM_001014288
|
Ptprd
|
protein tyrosine phosphatase, receptor type, D
|
|
chr5_-_143668403
|
3.721
|
|
Actb
|
actin, beta
|
|
chr5_+_91201538
|
3.673
|
|
Pf4
|
platelet factor 4
|
|
chr14_+_70857227
|
3.458
|
NM_145981
|
Phyhip
|
phytanoyl-CoA hydroxylase interacting protein
|
|
chr15_-_75950624
|
3.432
|
NM_144848
|
Eppk1
BC024139
|
epiplakin 1
cDNA sequence BC024139
|
|
chr14_+_21527175
|
3.427
|
|
2310021P13Rik
|
RIKEN cDNA 2310021P13 gene
|
|
chrX_-_97821244
|
3.410
|
NM_028303
|
Pdzd11
|
PDZ domain containing 11
|
|
chr2_-_119916315
|
3.379
|
|
Ehd4
|
EH-domain containing 4
|
|
chr4_+_148913978
|
3.366
|
|
Ctnnbip1
|
catenin beta interacting protein 1
|
|
chr9_+_53698053
|
3.292
|
NM_025540
|
Sln
|
sarcolipin
|
|
chr1_-_75250520
|
3.278
|
|
Ptprn
|
protein tyrosine phosphatase, receptor type, N
|
|
chr1_+_90003410
|
3.271
|
|
Ugt1a6b
|
UDP glucuronosyltransferase 1 family, polypeptide A6B
|
|
chr9_-_45012097
|
3.205
|
NM_145403
|
Tmprss4
|
transmembrane protease, serine 4
|
|
chr12_+_71008808
|
3.179
|
|
Atl1
|
atlastin GTPase 1
|
|
chr15_-_75901305
|
3.158
|
|
Puf60
|
poly-U binding splicing factor 60
|
|
chr15_-_101700048
|
3.134
|
NM_001003667
|
Krt77
|
keratin 77
|
|
chr5_+_73797316
|
3.118
|
NM_181323
|
Cwh43
|
cell wall biogenesis 43 C-terminal homolog (S. cerevisiae)
|
|
chr5_-_143668398
|
3.084
|
NM_007393
|
Actb
|
actin, beta
|
|
chr7_-_4474044
|
3.077
|
NM_009406
|
Tnni3
|
troponin I, cardiac 3
|
|
chr16_-_5222390
|
2.996
|
NM_001033220
|
AU021092
|
expressed sequence AU021092
|
|
chr5_-_124028994
|
2.963
|
|
Clip1
|
CAP-GLY domain containing linker protein 1
|
|
chr4_-_82151590
|
2.940
|
|
Nfib
|
nuclear factor I/B
|
|
chr9_+_15043658
|
2.911
|
|
2200002K05Rik
|
RIKEN cDNA 2200002K05 gene
|
|
chr19_+_34175422
|
2.902
|
NM_023903
|
Lipm
|
lipase, family member M
|
|
chr18_-_36886375
|
2.861
|
|
Cd14
|
CD14 antigen
|
|
chr19_-_6982516
|
2.771
|
|
Prdx5
|
peroxiredoxin 5
|
|
chr16_-_5222354
|
2.733
|
|
AU021092
|
expressed sequence AU021092
|
|
chr2_+_164388211
|
2.728
|
NM_026323
|
Wfdc2
|
WAP four-disulfide core domain 2
|
|
chr7_-_147298872
|
2.642
|
|
Echs1
|
enoyl Coenzyme A hydratase, short chain, 1, mitochondrial
|
|
chr10_+_96942124
|
2.599
|
NM_001190451
|
Dcn
|
decorin
|
|
chr12_-_4344470
|
2.594
|
|
Ncoa1
|
nuclear receptor coactivator 1
|
|
chr11_-_11832366
|
2.582
|
|
Grb10
|
growth factor receptor bound protein 10
|
|
chr17_+_35204268
|
2.580
|
NM_023463
|
Ly6g6c
|
lymphocyte antigen 6 complex, locus G6C
|
|
chr8_+_47576483
|
2.556
|
|
Acsl1
|
acyl-CoA synthetase long-chain family member 1
|
|
chr4_+_53453302
|
2.526
|
|
Slc44a1
|
solute carrier family 44, member 1
|
|
chr9_-_95745663
|
2.525
|
NM_001033210
|
Pls1
|
plastin 1 (I-isoform)
|
|
chr3_+_94497485
|
2.439
|
NM_019414
|
Selenbp2
|
selenium binding protein 2
|
|
chr7_-_51920870
|
2.373
|
NM_028021
|
Myh14
|
myosin, heavy polypeptide 14
|
|
chr2_+_143768541
|
2.356
|
|
Dstn
|
destrin
|
|
chr4_-_76240202
|
2.347
|
NM_011211
|
Ptprd
|
protein tyrosine phosphatase, receptor type, D
|
|
chr2_+_121134843
|
2.337
|
|
Mtap1a
|
microtubule-associated protein 1 A
|
|
chr18_-_36886307
|
2.280
|
NM_009841
|
Cd14
|
CD14 antigen
|
|
chr10_+_21698470
|
2.252
|
NM_001161847
|
Sgk1
|
serum/glucocorticoid regulated kinase 1
|
|
chr2_+_109514856
|
2.233
|
NM_007540
|
Bdnf
|
brain derived neurotrophic factor
|
|
chr15_-_89256262
|
2.232
|
NM_009948
|
Cpt1b
|
carnitine palmitoyltransferase 1b, muscle
|
|
chr1_-_166368439
|
2.167
|
|
Atp1b1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide
|
|
chr8_+_74092759
|
2.148
|
|
Slc27a1
|
solute carrier family 27 (fatty acid transporter), member 1
|
|
chr1_-_36756687
|
2.136
|
|
Actr1b
|
ARP1 actin-related protein 1 homolog B, centractin beta (yeast)
|
|
chr5_-_9725158
|
2.117
|
|
Grm3
|
glutamate receptor, metabotropic 3
|
|
chr6_+_22825464
|
2.063
|
NM_001081306
|
Ptprz1
|
protein tyrosine phosphatase, receptor type Z, polypeptide 1
|
|
chrX_+_133200913
|
2.052
|
NM_001029978
|
Tceal3
|
transcription elongation factor A (SII)-like 3
|
|
chr11_-_105805442
|
2.050
|
NM_007805
|
Cyb561
|
cytochrome b-561
|
|
chr5_-_51848420
|
2.019
|
|
Ppargc1a
|
peroxisome proliferative activated receptor, gamma, coactivator 1 alpha
|
|
chr10_+_56097078
|
1.981
|
NM_010288
|
Gja1
|
gap junction protein, alpha 1
|
|
chr8_+_74092790
|
1.968
|
NM_011977
|
Slc27a1
|
solute carrier family 27 (fatty acid transporter), member 1
|
|
chr15_-_89256252
|
1.945
|
|
Cpt1b
|
carnitine palmitoyltransferase 1b, muscle
|
|
chr19_-_59018112
|
1.929
|
|
Hspa12a
|
heat shock protein 12A
|
|
chr19_-_10379342
|
1.844
|
|
Dagla
|
diacylglycerol lipase, alpha
|
|
chr9_+_15043488
|
1.827
|
NM_026955
|
2200002K05Rik
|
RIKEN cDNA 2200002K05 gene
|
|
chr9_+_50302624
|
1.820
|
NM_029639
|
1600029D21Rik
|
RIKEN cDNA 1600029D21 gene
|
|
chr3_-_105746659
|
1.810
|
|
Atp5f1
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit B1
|
|
chr16_+_62814501
|
1.810
|
NM_026588
|
Stx19
|
syntaxin 19
|
|
chr2_+_135567565
|
1.804
|
NM_013829
|
Plcb4
|
phospholipase C, beta 4
|
|
chr14_+_80400057
|
1.796
|
NM_001030294
|
Olfm4
|
olfactomedin 4
|
|
chr11_-_83881811
|
1.794
|
NM_019819
|
Dusp14
|
dual specificity phosphatase 14
|
|
chr11_-_70135960
|
1.779
|
NM_145684
|
Alox12e
|
arachidonate lipoxygenase, epidermal
|
|
chr18_+_38456315
|
1.767
|
|
Rnf14
|
ring finger protein 14
|
|
chr17_-_90436488
|
1.759
|
|
Nrxn1
|
neurexin I
|
|
chr1_+_151947225
|
1.748
|
NM_011198
|
Ptgs2
|
prostaglandin-endoperoxide synthase 2
|
|
chr3_-_107820705
|
1.720
|
|
Gstm1
|
glutathione S-transferase, mu 1
|
|
chr9_-_14916551
|
1.689
|
NM_001164797
|
Hephl1
|
hephaestin-like 1
|
|
chr19_-_10379361
|
1.680
|
NM_198114
|
Dagla
|
diacylglycerol lipase, alpha
|
|
chr11_-_106163120
|
1.677
|
NM_008117
|
Gh
|
growth hormone
|
|
chr11_-_59025422
|
1.630
|
|
Arf1
|
ADP-ribosylation factor 1
|
|
chr7_-_31911733
|
1.625
|
|
Scn1b
|
sodium channel, voltage-gated, type I, beta
|
|
chr11_-_69499014
|
1.613
|
NM_001159505
NM_023517
|
Tnfsf13
|
tumor necrosis factor (ligand) superfamily, member 13
|
|
chr2_+_138104219
|
1.591
|
NM_145534
|
Btbd3
|
BTB (POZ) domain containing 3
|
|
chr10_+_115614313
|
1.580
|
NM_001161839
NM_001161838
NM_001161840
|
Ptprr
|
protein tyrosine phosphatase, receptor type, R
|
|
chr7_+_17334480
|
1.533
|
|
Ap2s1
|
adaptor-related protein complex 2, sigma 1 subunit
|
|
chr11_+_28753190
|
1.452
|
NM_146015
|
Efemp1
|
epidermal growth factor-containing fibulin-like extracellular matrix protein 1
|
|
chr11_-_83880574
|
1.439
|
|
Dusp14
|
dual specificity phosphatase 14
|
|
chr3_+_37538808
|
1.423
|
NM_011896
|
Spry1
|
sprouty homolog 1 (Drosophila)
|
|
chr2_-_80421054
|
1.397
|
NM_016965
|
Nckap1
|
NCK-associated protein 1
|
|
chr6_+_57653067
|
1.393
|
|
Lancl2
|
LanC (bacterial lantibiotic synthetase component C)-like 2
|
|
chr1_+_74283325
|
1.369
|
|
Arpc2
|
actin related protein 2/3 complex, subunit 2
|
|
chr17_-_24306262
|
1.354
|
NM_009729
|
Atp6v0c-ps2
Atp6v0c
|
ATPase, H+ transporting, lysosomal V0 subunit C, pseudogene 2
ATPase, H+ transporting, lysosomal V0 subunit C
|
|
chrX_+_38754480
|
1.347
|
NM_016886
|
Gria3
|
glutamate receptor, ionotropic, AMPA3 (alpha 3)
|
|
chr3_-_97628441
|
1.308
|
NM_001110163
|
Pde4dip
|
phosphodiesterase 4D interacting protein (myomegalin)
|
|
chr11_-_99982369
|
1.301
|
|
Krt13
|
keratin 13
|
|
chr13_-_74480876
|
1.273
|
|
Sdha
|
succinate dehydrogenase complex, subunit A, flavoprotein (Fp)
|
|
chr1_+_90034940
|
1.238
|
|
Ugt1a6a
|
UDP glucuronosyltransferase 1 family, polypeptide A6A
|
|
chr14_+_32064648
|
1.238
|
NM_027088
|
Bap1
|
Brca1 associated protein 1
|
|
chr3_-_92655207
|
1.233
|
NM_028628
|
Lce1l
|
late cornified envelope 1L
|
|
chr3_-_105746740
|
1.231
|
|
Atp5f1
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit B1
|
|
chr2_+_25312352
|
1.224
|
|
Clic3
|
chloride intracellular channel 3
|
|
chr1_-_164816645
|
1.220
|
|
Fmo2
|
flavin containing monooxygenase 2
|
|
chr11_-_94254221
|
1.219
|
NM_029600
|
Abcc3
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 3
|
|
chr2_+_160706405
|
1.219
|
NM_022883
|
Lpin3
|
lipin 3
|
|
chr9_-_99610481
|
1.190
|
NM_001194922
NM_019815
|
Cldn18
|
claudin 18
|
|
chr18_+_45719464
|
1.169
|
|
Kcnn2
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 2
|
|
chr5_-_144266956
|
1.165
|
|
|
|
|
chr11_+_68933954
|
1.157
|
NM_033041
|
Hes7
|
hairy and enhancer of split 7 (Drosophila)
|
|
chr4_+_12833983
|
1.144
|
NM_001171801
NM_173746
|
Gm11818
|
predicted gene 11818
|
|
chr14_-_66539632
|
1.141
|
|
Scara3
|
scavenger receptor class A, member 3
|
|
chr2_-_164008333
|
1.129
|
NM_145369
|
Wfdc5
|
WAP four-disulfide core domain 5
|
|
chr18_+_38456280
|
1.108
|
NM_001164622
NM_020012
|
Rnf14
|
ring finger protein 14
|
|
chr18_+_38456364
|
1.105
|
NM_001164621
|
Rnf14
|
ring finger protein 14
|
|
chr8_+_11834520
|
1.103
|
|
Arhgef7
|
Rho guanine nucleotide exchange factor (GEF7)
|
|
chr10_+_87922331
|
1.101
|
NM_011517
|
Sycp3
|
synaptonemal complex protein 3
|
|
chr2_+_28368603
|
1.098
|
NM_001145834
NM_009058
|
Ralgds
|
ral guanine nucleotide dissociation stimulator
|
|
chrX_+_130829763
|
1.096
|
NM_026239
|
Tmem35
|
transmembrane protein 35
|
|
chr5_-_23897960
|
1.085
|
NM_001002897
|
Atg9b
|
ATG9 autophagy related 9 homolog B (S. cerevisiae)
|
|
chr10_+_98453513
|
1.080
|
|
Atp2b1
|
ATPase, Ca++ transporting, plasma membrane 1
|
|
chr15_-_83002561
|
1.077
|
|
Cyb5r3
|
cytochrome b5 reductase 3
|
|
chr18_-_39078304
|
1.070
|
NM_010197
|
Fgf1
|
fibroblast growth factor 1
|
|
chr18_-_36886051
|
1.064
|
|
Cd14
|
CD14 antigen
|
|
chr14_-_100548982
|
1.061
|
NM_010636
|
Klf12
|
Kruppel-like factor 12
|
|
chr16_+_94664089
|
1.057
|
|
Ttc3
|
tetratricopeptide repeat domain 3
|
|
chr8_+_131238027
|
1.054
|
|
Itgb1
|
integrin beta 1 (fibronectin receptor beta)
|
|
chrX_-_130432840
|
1.045
|
NM_019656
|
Tspan6
|
tetraspanin 6
|
|
chr14_+_55502261
|
1.044
|
NM_007537
|
Bcl2l2
|
BCL2-like 2
|
|
chrX_+_97925185
|
1.043
|
NM_023608
|
Gdpd2
|
glycerophosphodiester phosphodiesterase domain containing 2
|
|
chr2_-_6110280
|
1.019
|
|
Echdc3
|
enoyl Coenzyme A hydratase domain containing 3
|
|
chr7_+_150293158
|
0.997
|
NM_008434
|
Kcnq1
|
potassium voltage-gated channel, subfamily Q, member 1
|
|
chr7_-_4116102
|
0.991
|
|
Cdc42ep5
|
CDC42 effector protein (Rho GTPase binding) 5
|
|
chr19_-_5844533
|
0.988
|
|
Neat1
|
nuclear paraspeckle assembly transcript 1 (non-protein coding)
|
|
chr8_+_24145593
|
0.977
|
|
Ank1
|
ankyrin 1, erythroid
|
|
chr3_-_89083562
|
0.971
|
NM_010107
|
Efna1
|
ephrin A1
|
|
chr5_-_31395853
|
0.965
|
NM_011773
|
Slc30a3
|
solute carrier family 30 (zinc transporter), member 3
|
|
chr9_+_54442900
|
0.963
|
|
Idh3a
|
isocitrate dehydrogenase 3 (NAD+) alpha
|
|
chr15_-_75397601
|
0.939
|
NM_011837
|
Ly6h
|
lymphocyte antigen 6 complex, locus H
|
|
chr2_+_143741243
|
0.938
|
|
Dstn
|
destrin
|
|
chr7_-_4473916
|
0.936
|
|
Tnni3
|
troponin I, cardiac 3
|
|
chr2_-_66278870
|
0.935
|
NM_018733
|
Scn1a
|
sodium channel, voltage-gated, type I, alpha
|
|
chr9_-_108469265
|
0.933
|
|
Ndufaf3
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, assembly factor 3
|
|
chr14_+_55502200
|
0.904
|
|
Bcl2l2
|
BCL2-like 2
|
|
chr3_-_92819586
|
0.897
|
NM_028798
|
Crct1
|
cysteine-rich C-terminal 1
|
|
chr2_+_3035629
|
0.873
|
NM_001081161
|
Fam171a1
|
family with sequence similarity 171, member A1
|
|
chr9_+_107483009
|
0.863
|
|
Nat6
|
N-acetyltransferase 6
|
|
chr5_-_109027924
|
0.859
|
|
Gak
|
cyclin G associated kinase
|
|
chr10_+_94039007
|
0.859
|
NM_177026
|
Tmcc3
|
transmembrane and coiled coil domains 3
|
|
chr6_-_139989932
|
0.845
|
NM_054066
|
Plcz1
|
phospholipase C, zeta 1
|
|
chr3_+_51028714
|
0.841
|
|
Ccrn4l
|
CCR4 carbon catabolite repression 4-like (S. cerevisiae)
|
|
chr5_+_114222723
|
0.836
|
NM_025526
|
Iscu
|
IscU iron-sulfur cluster scaffold homolog (E. coli)
|
|
chr7_-_139749889
|
0.836
|
|
|
|
|
chr2_-_110203298
|
0.832
|
|
Fibin
|
fin bud initiation factor homolog (zebrafish)
|
|
chrX_+_162782484
|
0.815
|
NM_001177955
NM_001177956
NM_001177957
NM_001177958
NM_001177959
NM_001177960
|
Gpm6b
|
glycoprotein m6b
|
|
chr2_-_12933305
|
0.810
|
|
C1ql3
|
C1q-like 3
|
|
chr6_+_114081227
|
0.787
|
NM_172890
|
Slc6a11
|
solute carrier family 6 (neurotransmitter transporter, GABA), member 11
|
|
chrX_+_38754568
|
0.767
|
|
Gria3
|
glutamate receptor, ionotropic, AMPA3 (alpha 3)
|
|
chr4_-_155574875
|
0.764
|
NM_015783
|
Isg15
|
ISG15 ubiquitin-like modifier
|
|
chr15_-_83002612
|
0.745
|
|
Cyb5r3
|
cytochrome b5 reductase 3
|
|
chr2_-_110203264
|
0.742
|
|
Fibin
|
fin bud initiation factor homolog (zebrafish)
|
|
chr18_-_12464213
|
0.741
|
NM_001190371
|
Ankrd29
|
ankyrin repeat domain 29
|
|
chr3_-_88307198
|
0.736
|
|
Lmna
|
lamin A
|
|
chr6_+_56828743
|
0.732
|
|
Fkbp9
|
FK506 binding protein 9
|
|
chr2_+_25312358
|
0.732
|
NM_027085
|
Clic3
|
chloride intracellular channel 3
|
|
chr11_+_66984528
|
0.720
|
NM_001039545
|
Myh2
|
myosin, heavy polypeptide 2, skeletal muscle, adult
|
|
chr19_-_6982094
|
0.719
|
|
Prdx5
|
peroxiredoxin 5
|
|
chr2_+_3035692
|
0.715
|
|
Fam171a1
|
family with sequence similarity 171, member A1
|
|
chr10_+_79608206
|
0.714
|
|
|
|
|
chr14_+_32064678
|
0.714
|
|
Bap1
|
Brca1 associated protein 1
|
|
chr3_-_10208480
|
0.708
|
|
Fabp4
Fabp9
|
fatty acid binding protein 4, adipocyte
fatty acid binding protein 9, testis
|
|
chr6_-_13789847
|
0.707
|
NM_145066
|
Gpr85
|
G protein-coupled receptor 85
|
|
chr1_+_169528706
|
0.691
|
|
Rxrg
|
retinoid X receptor gamma
|
|
chr6_+_7505070
|
0.674
|
NM_009311
|
Tac1
|
tachykinin 1
|
|
chr2_-_32136262
|
0.672
|
|
|
|
|
chr11_+_93905557
|
0.665
|
|
Spag9
|
sperm associated antigen 9
|