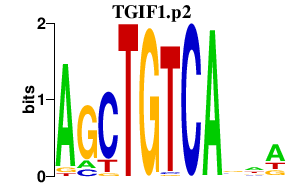
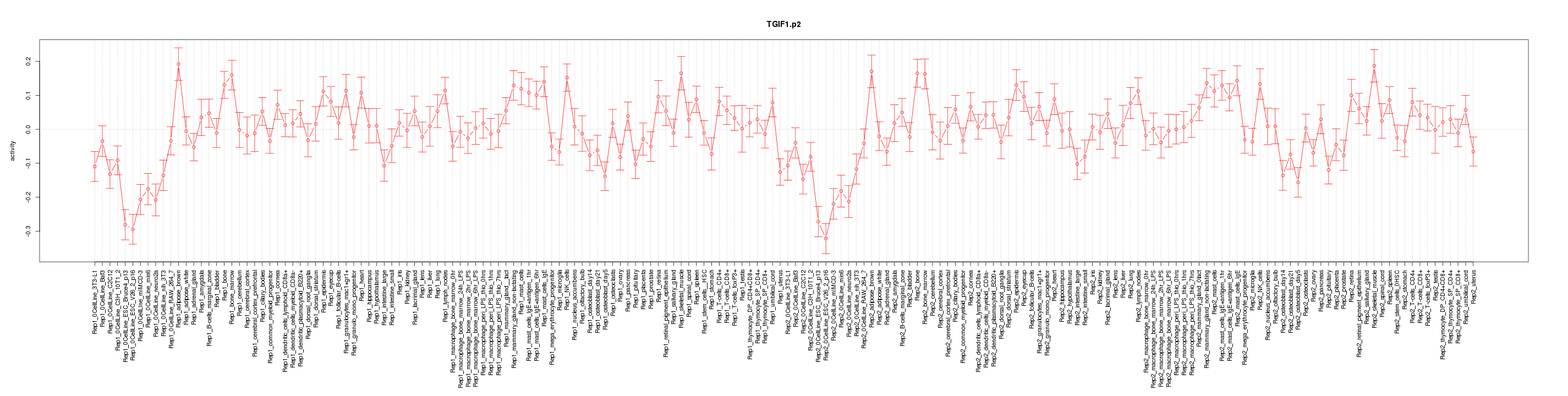
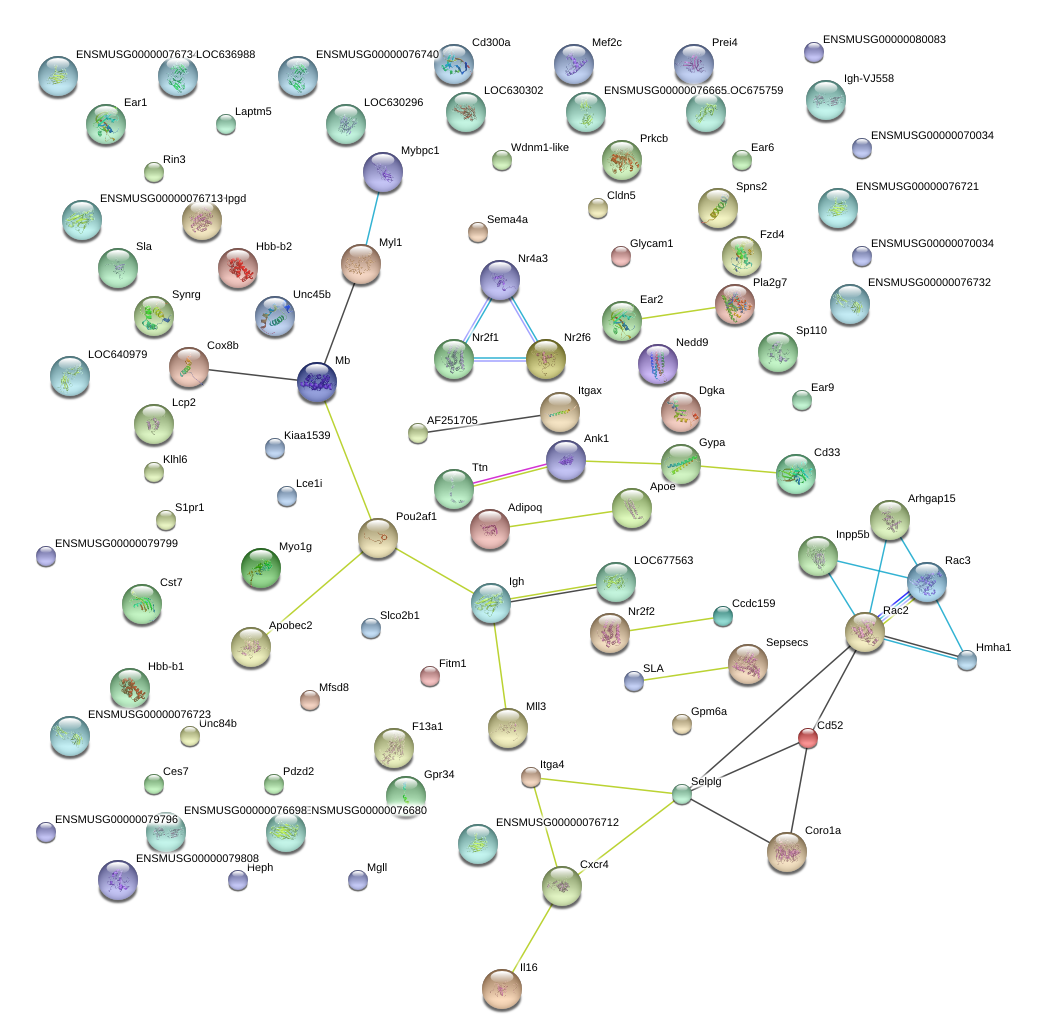
|
chr5_-_114280434
|
20.752
|
NM_009151
|
Selplg
|
selectin, platelet (p-selectin) ligand
|
|
chr7_-_110962416
|
18.964
|
NM_008220
NM_016956
|
Hbb-b1
Hbb-b2
|
hemoglobin, beta adult major chain
hemoglobin, beta adult minor chain
|
|
chr16_-_19983049
|
18.732
|
NM_183390
|
Klhl6
|
kelch-like 6 (Drosophila)
|
|
chr7_-_110976438
|
17.539
|
|
Hbb-b1
|
hemoglobin, beta adult major chain
|
|
chr7_-_110976439
|
16.855
|
|
Hbb-b1
|
hemoglobin, beta adult major chain
|
|
chr15_-_78403110
|
16.727
|
NM_009008
|
Rac2
|
RAS-related C3 botulinum substrate 2
|
|
chr13_+_83643032
|
15.191
|
NM_001170537
NM_025282
|
Mef2c
|
myocyte enhancer factor 2C
|
|
chr1_-_182126032
|
15.018
|
NM_023341
|
Adck3
|
aarF domain containing kinase 3
|
|
chr8_+_58773380
|
13.965
|
|
Hpgd
|
hydroxyprostaglandin dehydrogenase 15 (NAD)
|
|
chr7_-_110976499
|
13.903
|
|
Hbb-b1
|
hemoglobin, beta adult major chain
|
|
chr8_+_58773321
|
13.844
|
NM_008278
|
Hpgd
|
hydroxyprostaglandin dehydrogenase 15 (NAD)
|
|
chr7_-_110962324
|
13.647
|
|
Hbb-b1
Hbb-b2
|
hemoglobin, beta adult major chain
hemoglobin, beta adult minor chain
|
|
chr4_+_130469048
|
13.482
|
|
Laptm5
|
lysosomal-associated protein transmembrane 5
|
|
chr4_+_48058102
|
13.316
|
|
Nr4a3
|
nuclear receptor subfamily 4, group A, member 3
|
|
chr1_-_66991625
|
12.648
|
NM_021285
|
Myl1
|
myosin, light polypeptide 1
|
|
chr4_+_130469187
|
12.523
|
NM_010686
|
Laptm5
|
lysosomal-associated protein transmembrane 5
|
|
chr8_+_83017933
|
12.199
|
NM_010369
|
Gypa
|
glycophorin A
|
|
chr14_+_56194510
|
12.120
|
NM_026808
|
Fitm1
|
fat storage-inducing transmembrane protein 1
|
|
chr7_-_106859849
|
11.851
|
NM_175316
|
Slco2b1
|
solute carrier organic anion transporter family, member 2b1
|
|
chr15_-_76853069
|
11.809
|
NM_001164048
NM_001164047
|
Mb
|
myoglobin
|
|
chr14_+_52473672
|
11.708
|
NM_017385
|
Ear7
|
eosinophil-associated, ribonuclease A family, member 7
|
|
chr6_+_88674821
|
11.546
|
|
Mgll
|
monoglyceride lipase
|
|
chr1_-_66991580
|
11.520
|
|
Myl1
|
myosin, light polypeptide 1
|
|
chr11_+_33946927
|
11.436
|
|
Lcp2
|
lymphocyte cytosolic protein 2
|
|
chr14_+_44680318
|
11.399
|
NM_007895
NM_001012766
NM_017388
|
BC151093
Ear2
Ear12
Ear3
|
cDNA sequence BC151093
eosinophil-associated, ribonuclease A family, member 2
eosinophil-associated, ribonuclease A family, member 12
eosinophil-associated, ribonuclease A family, member 3
|
|
chr11_-_72303447
|
11.379
|
|
Spns2
|
spinster homolog 2 (Drosophila)
|
|
chr17_-_48572043
|
11.229
|
NM_009694
|
Apobec2
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 2
|
|
chr17_-_48571942
|
11.116
|
|
Apobec2
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 2
|
|
chr8_+_24249481
|
10.970
|
|
Ank1
|
ankyrin 1, erythroid
|
|
chr7_-_110976314
|
10.937
|
|
Hbb-b1
Hbb-b2
|
hemoglobin, beta adult major chain
hemoglobin, beta adult minor chain
|
|
chr2_-_76820599
|
10.916
|
NM_011652
NM_028004
|
Ttn
|
titin
|
|
chr15_-_66644127
|
10.751
|
NM_009192
|
Sla
|
src-like adaptor
|
|
chr11_+_83560437
|
10.542
|
NM_183249
|
1100001G20Rik
|
RIKEN cDNA 1100001G20 gene
|
|
chr2_+_150396150
|
10.500
|
NM_009977
|
Cst7
|
cystatin F (leukocystatin)
|
|
chr7_+_129432585
|
10.474
|
NM_008855
|
Prkcb
|
protein kinase C, beta
|
|
chr7_-_90883999
|
10.464
|
NM_010551
|
Il16
|
interleukin 16
|
|
chr2_-_132403964
|
10.427
|
NM_001042671
NM_001042672
NM_028802
|
Gpcpd1
|
glycerophosphocholine phosphodiesterase GDE1 homolog (S. cerevisiae)
|
|
chr2_-_132403832
|
10.261
|
|
Gpcpd1
|
glycerophosphocholine phosphodiesterase GDE1 homolog (S. cerevisiae)
|
|
chr6_+_88674398
|
10.256
|
NM_001166249
NM_011844
|
Mgll
|
monoglyceride lipase
|
|
chr7_-_20283542
|
10.081
|
|
Apoe
|
apolipoprotein E
|
|
chr4_-_133650937
|
10.058
|
NM_013706
|
Cd52
|
CD52 antigen
|
|
chr11_-_6420904
|
9.971
|
|
Myo1g
|
myosin IG
|
|
chr3_-_88259594
|
9.920
|
|
Sema4a
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A
|
|
chr11_+_114751585
|
9.837
|
|
Cd300a
|
CD300A antigen
|
|
chr6_+_68375560
|
9.761
|
|
Igkv16-104
|
immunoglobulin kappa variable 16-104
|
|
chr16_+_18777032
|
9.657
|
|
Cldn5
|
claudin 5
|
|
chr7_-_133848235
|
9.618
|
NM_009898
|
Coro1a
|
coronin, actin binding protein 1A
|
|
chr11_-_72303293
|
9.611
|
NM_153060
|
Spns2
|
spinster homolog 2 (Drosophila)
|
|
chr14_-_44501032
|
9.540
|
NM_053112
|
Ear10
|
eosinophil-associated, ribonuclease A family, member 10
|
|
chr10_+_79479416
|
9.441
|
NM_001142701
|
Hmha1
|
histocompatibility (minor) HA-1
|
|
chr3_-_88259621
|
9.119
|
|
Sema4a
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A
|
|
chr7_-_50788445
|
9.053
|
|
Cd33
|
CD33 antigen
|
|
chr3_-_88259682
|
9.016
|
NM_013658
|
Sema4a
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A
|
|
chr17_+_43705510
|
8.985
|
|
Pla2g7
|
phospholipase A2, group VII (platelet-activating factor acetylhydrolase, plasma)
|
|
chr15_-_66644199
|
8.872
|
|
Sla
|
src-like adaptor
|
|
chr5_-_25004519
|
8.809
|
|
Mll3
|
myeloid/lymphoid or mixed-lineage leukemia 3
|
|
chr7_-_50788540
|
8.686
|
NM_001111058
NM_021293
|
Cd33
|
CD33 antigen
|
|
chr13_+_83803830
|
8.427
|
|
Mef2c
|
myocyte enhancer factor 2C
|
|
chr7_+_135273060
|
8.413
|
NM_021334
|
Itgax
|
integrin alpha X
|
|
chr3_+_17695743
|
8.408
|
|
6430547I21Rik
|
RIKEN cDNA 6430547I21 gene
|
|
chr13_-_37141055
|
8.381
|
NM_028784
|
F13a1
|
coagulation factor XIII, A1 subunit
|
|
chr1_-_130488785
|
8.348
|
NM_009911
|
Cxcr4
|
chemokine (C-X-C motif) receptor 4
|
|
chr11_+_82724754
|
8.220
|
NM_178680
|
Unc45b
|
unc-45 homolog B (C. elegans)
|
|
chr16_+_23146631
|
8.169
|
|
Adipoq
|
adiponectin, C1Q and collagen domain containing
|
|
chr11_+_83777922
|
8.134
|
NM_001115009
NM_194341
|
Synrg
|
synergin, gamma
|
|
chr3_-_115417847
|
8.091
|
|
S1pr1
|
sphingosine-1-phosphate receptor 1
|
|
chr10_-_128181069
|
8.066
|
NM_016811
|
Dgka
|
diacylglycerol kinase, alpha
|
|
chr10_-_88067758
|
8.056
|
NM_175418
|
Mybpc1
|
myosin binding protein C, slow-type
|
|
chr15_-_79572885
|
8.050
|
NM_194342
|
Sun2
|
Sad1 and UNC84 domain containing 2
|
|
chr16_+_18776936
|
8.030
|
NM_013805
|
Cldn5
|
claudin 5
|
|
chr7_+_96552955
|
7.946
|
|
Fzd4
|
frizzled homolog 4 (Drosophila)
|
|
chr7_-_148086285
|
7.940
|
|
Cox8b
|
cytochrome c oxidase, subunit VIIIb
|
|
chr16_+_23146608
|
7.926
|
NM_009605
|
Adipoq
|
adiponectin, C1Q and collagen domain containing
|
|
chr14_+_56194622
|
7.899
|
|
Fitm1
|
fat storage-inducing transmembrane protein 1
|
|
chr15_-_103395483
|
7.866
|
NM_008134
|
Glycam1
|
glycosylation dependent cell adhesion molecule 1
|
|
chr1_-_87500465
|
7.863
|
NM_175397
|
Sp110
|
Sp110 nuclear body protein
|
|
chr12_+_103521355
|
7.847
|
|
Rin3
|
Ras and Rab interactor 3
|
|
chr16_+_18776955
|
7.832
|
|
Cldn5
|
claudin 5
|
|
chr8_+_56040553
|
7.796
|
|
Gpm6a
|
glycoprotein m6a
|
|
chr4_+_124419115
|
7.778
|
NM_008385
|
Inpp5b
|
inositol polyphosphate-5-phosphatase B
|
|
chrX_+_13209894
|
7.774
|
NM_011823
|
Gpr34
|
G protein-coupled receptor 34
|
|
chr9_+_51021793
|
7.740
|
NM_011136
|
Pou2af1
|
POU domain, class 2, associating factor 1
|
|
chr7_-_80703275
|
7.702
|
|
1810026B05Rik
|
RIKEN cDNA 1810026B05 gene
|
|
chr4_-_43059017
|
7.605
|
NM_172691
|
B230312A22Rik
|
RIKEN cDNA B230312A22 gene
|
|
chr2_+_79095582
|
7.566
|
NM_010576
|
Itga4
|
integrin alpha 4
|
|
chr2_+_43604343
|
7.460
|
NM_153820
NM_001025377
|
Arhgap15
|
Rho GTPase activating protein 15
|
|
chr13_-_41454358
|
7.399
|
NM_017464
|
Nedd9
|
neural precursor cell expressed, developmentally down-regulated gene 9
|
|
chr3_-_40650624
|
7.316
|
NM_028140
|
Mfsd8
|
major facilitator superfamily domain containing 8
|
|
chr12_-_115526405
|
7.106
|
|
Igh-VJ558
Igh-2
LOC432699
|
immunoglobulin heavy chain (J558 family)
immunoglobulin heavy chain 2 (serum IgA)
VH gene product
|
|
chr3_-_115417932
|
7.091
|
NM_007901
|
S1pr1
|
sphingosine-1-phosphate receptor 1
|
|
chr3_-_92582820
|
7.075
|
NM_029667
|
Lce1i
|
late cornified envelope 1I
|
|
chr15_-_12479631
|
7.066
|
|
Pdzd2
|
PDZ domain containing 2
|
|
chr12_+_103521251
|
7.048
|
NM_177620
|
Rin3
|
Ras and Rab interactor 3
|
|
chr15_-_41700964
|
6.997
|
NM_175456
|
Abra
|
actin-binding Rho activating protein
|
|
chr6_+_97757012
|
6.981
|
NM_001113198
|
Mitf
|
microphthalmia-associated transcription factor
|
|
chr15_+_81633102
|
6.948
|
NM_153484
|
Tef
|
thyrotroph embryonic factor
|
|
chr15_+_82129383
|
6.938
|
NM_029066
|
Wbp2nl
|
WBP2 N-terminal like
|
|
chr10_+_41530379
|
6.867
|
NM_001162908
|
Sesn1
|
sestrin 1
|
|
chr4_-_43058685
|
6.843
|
|
B230312A22Rik
|
RIKEN cDNA B230312A22 gene
|
|
chr12_+_103521406
|
6.783
|
|
Rin3
|
Ras and Rab interactor 3
|
|
chr17_+_87153083
|
6.722
|
NM_010137
|
Epas1
|
endothelial PAS domain protein 1
|
|
chr7_-_139978601
|
6.699
|
NM_175268
|
Fam53b
|
family with sequence similarity 53, member B
|
|
chr1_+_94807324
|
6.673
|
|
Rnpepl1
|
arginyl aminopeptidase (aminopeptidase B)-like 1
|
|
chr8_+_56040445
|
6.667
|
|
Gpm6a
|
glycoprotein m6a
|
|
chr16_-_46120221
|
6.621
|
NM_032465
|
Cd96
|
CD96 antigen
|
|
chr11_+_78640059
|
6.560
|
NM_001076681
|
1810012P15Rik
|
RIKEN cDNA 1810012P15 gene
|
|
chr17_+_35516558
|
6.482
|
NM_001143689
|
H2-Gs10
|
MHC class I like protein GS10
|
|
chr3_-_115417685
|
6.461
|
|
S1pr1
|
sphingosine-1-phosphate receptor 1
|
|
chr11_+_58020859
|
6.419
|
|
Igtp
|
interferon gamma induced GTPase
|
|
chr3_-_112959644
|
6.360
|
NM_001190403
NM_001190404
|
Amy2b
|
amylase 2b
|
|
chr15_-_5058532
|
6.337
|
NM_001163138
|
Card6
|
caspase recruitment domain family, member 6
|
|
chr11_+_100442831
|
6.239
|
|
Cnp
|
2',3'-cyclic nucleotide 3' phosphodiesterase
|
|
chr7_+_96552871
|
6.214
|
NM_008055
|
Fzd4
|
frizzled homolog 4 (Drosophila)
|
|
chr4_+_140977613
|
6.072
|
|
Hspb7
|
heat shock protein family, member 7 (cardiovascular)
|
|
chr14_-_44397257
|
6.026
|
NM_007894
|
Ear1
|
eosinophil-associated, ribonuclease A family, member 1
|
|
chr14_+_56194658
|
6.004
|
|
Fitm1
|
fat storage-inducing transmembrane protein 1
|
|
chr17_+_75835208
|
6.004
|
NM_001166493
|
Rasgrp3
|
RAS, guanyl releasing protein 3
|
|
chr2_+_69508174
|
6.002
|
NM_001081087
|
Kbtbd10
|
kelch repeat and BTB (POZ) domain containing 10
|
|
chr17_-_37712365
|
5.980
|
NM_146289
|
Olfr113
|
olfactory receptor 113
|
|
chr16_-_89818596
|
5.972
|
NM_001145887
|
Tiam1
|
T-cell lymphoma invasion and metastasis 1
|
|
chr6_+_48689045
|
5.914
|
NM_008376
NM_175860
|
Gimap1
|
GTPase, IMAP family member 1
|
|
chr8_+_56040080
|
5.813
|
NM_153581
|
Gpm6a
|
glycoprotein m6a
|
|
chr6_+_50060895
|
5.740
|
|
Mpp6
|
membrane protein, palmitoylated 6 (MAGUK p55 subfamily member 6)
|
|
chr3_-_113094211
|
5.733
|
NM_001160150
NM_001160151
NM_001160152
NM_001042711
|
Amy2a4
Amy2a3
Amy2a2
Amy2a5
|
amylase 2a4
amylase 2a3
amylase 2a2
amylase 2a5
|
|
chr5_-_44617844
|
5.733
|
NM_173764
|
Tapt1
|
transmembrane anterior posterior transformation 1
|
|
chr11_+_78640103
|
5.730
|
|
1810012P15Rik
|
RIKEN cDNA 1810012P15 gene
|
|
chr17_-_27304602
|
5.725
|
NM_173027
|
Ip6k3
|
inositol hexaphosphate kinase 3
|
|
chr8_+_122638031
|
5.717
|
NM_001163761
NM_001163762
NM_172286
|
6430548M08Rik
|
RIKEN cDNA 6430548M08 gene
|
|
chr3_-_116671573
|
5.695
|
|
Palmd
|
palmdelphin
|
|
chr7_-_90803895
|
5.632
|
|
Il16
|
interleukin 16
|
|
chr14_-_73725546
|
5.565
|
|
Rb1
|
retinoblastoma 1
|
|
chr4_-_22415444
|
5.451
|
|
Pou3f2
|
POU domain, class 3, transcription factor 2
|
|
chr2_-_160192737
|
5.439
|
|
Mafb
|
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein B (avian)
|
|
chr14_+_56179739
|
5.437
|
NM_001199009
|
Dcaf11
|
DDB1 and CUL4 associated factor 11
|
|
chr11_+_33947115
|
5.418
|
NM_010696
|
Lcp2
|
lymphocyte cytosolic protein 2
|
|
chr1_+_137949360
|
5.414
|
NM_001081023
NM_014193
|
Cacna1s
|
calcium channel, voltage-dependent, L type, alpha 1S subunit
|
|
chr2_-_113878535
|
5.379
|
NM_009608
|
Actc1
|
actin, alpha, cardiac muscle 1
|
|
chr6_+_55286292
|
5.349
|
NM_007472
|
Aqp1
|
aquaporin 1
|
|
chr5_-_25004598
|
5.337
|
NM_001081383
|
Mll3
|
myeloid/lymphoid or mixed-lineage leukemia 3
|
|
chr12_+_113919213
|
5.294
|
|
Zbtb42
|
zinc finger and BTB domain containing 42
|
|
chrX_+_154137193
|
5.286
|
|
Smpx
|
small muscle protein, X-linked
|
|
chr8_-_63461923
|
5.253
|
|
Clcn3
|
chloride channel 3
|
|
chr11_-_108205199
|
5.242
|
NM_011101
|
Prkca
|
protein kinase C, alpha
|
|
chr10_+_115738429
|
5.225
|
NM_029928
|
Ptprb
|
protein tyrosine phosphatase, receptor type, B
|
|
chr17_+_35561811
|
5.216
|
NM_207648
|
H2-Q6
|
histocompatibility 2, Q region locus 6
|
|
chr11_+_82930294
|
5.175
|
NM_011407
|
Slfn1
|
schlafen 1
|
|
chr10_-_80131318
|
5.169
|
|
Mknk2
|
MAP kinase-interacting serine/threonine kinase 2
|
|
chr8_+_47556503
|
5.147
|
|
Acsl1
|
acyl-CoA synthetase long-chain family member 1
|
|
chr17_+_43705388
|
5.136
|
NM_013737
|
Pla2g7
|
phospholipase A2, group VII (platelet-activating factor acetylhydrolase, plasma)
|
|
chr7_+_63023547
|
5.090
|
NM_178706
|
Siglech
|
sialic acid binding Ig-like lectin H
|
|
chr7_+_29225955
|
5.083
|
NM_001039192
|
Gmfg
|
glia maturation factor, gamma
|
|
chr1_+_155747072
|
5.067
|
NM_008131
|
Glul
|
glutamate-ammonia ligase (glutamine synthetase)
|
|
chr17_+_35607032
|
5.066
|
NM_010391
|
H2-Q10
|
histocompatibility 2, Q region locus 10
|
|
chr7_+_138554108
|
5.064
|
NM_025826
|
Acadsb
|
acyl-Coenzyme A dehydrogenase, short/branched chain
|
|
chr11_-_86539846
|
5.051
|
|
Cltc
|
clathrin, heavy polypeptide (Hc)
|
|
chr12_+_111723922
|
5.045
|
NM_001081457
NM_001081458
NM_012023
|
Ppp2r5c
|
protein phosphatase 2, regulatory subunit B (B56), gamma isoform
|
|
chr4_+_140977621
|
5.008
|
|
Hspb7
|
heat shock protein family, member 7 (cardiovascular)
|
|
chr17_+_35400056
|
5.004
|
|
H2-D1
H2-L
|
histocompatibility 2, D region locus 1
histocompatibility 2, D region
|
|
chr7_+_138554180
|
4.959
|
|
Acadsb
|
acyl-Coenzyme A dehydrogenase, short/branched chain
|
|
chr4_+_147374850
|
4.948
|
NM_008725
|
Nppa
|
natriuretic peptide type A
|
|
chr3_-_113277601
|
4.947
|
NM_007446
|
Amy1
|
amylase 1, salivary
|
|
chr2_-_11423651
|
4.922
|
|
Pfkfb3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3
|
|
chr3_-_144382959
|
4.912
|
NM_019464
|
Sh3glb1
|
SH3-domain GRB2-like B1 (endophilin)
|
|
chr5_+_137729501
|
4.885
|
NM_009599
|
Ache
|
acetylcholinesterase
|
|
chr15_+_81297235
|
4.876
|
|
Rbx1
|
ring-box 1
|
|
chrX_+_154137049
|
4.863
|
NM_025357
|
Smpx
|
small muscle protein, X-linked
|
|
chr13_+_24706474
|
4.837
|
NM_178658
|
Fam65b
|
family with sequence similarity 65, member B
|
|
chrX_+_137491455
|
4.824
|
NM_174875
|
Atg4a
|
autophagy-related 4A (yeast)
|
|
chr6_+_49269294
|
4.803
|
|
Ccdc126
|
coiled-coil domain containing 126
|
|
chr9_+_64659238
|
4.792
|
|
Dennd4a
|
DENN/MADD domain containing 4A
|
|
chr6_-_106750030
|
4.780
|
NM_021449
NM_175357
|
Crbn
|
cereblon
|
|
chrX_+_154137114
|
4.772
|
|
Smpx
|
small muscle protein, X-linked
|
|
chr11_+_114751354
|
4.731
|
NM_170758
|
Cd300a
|
CD300A antigen
|
|
chr11_-_96976562
|
4.700
|
NM_019507
|
Tbx21
|
T-box 21
|
|
chr8_+_56040211
|
4.691
|
|
Gpm6a
|
glycoprotein m6a
|
|
chr17_+_44325518
|
4.690
|
NM_172621
|
Clic5
|
chloride intracellular channel 5
|
|
chr10_+_122116066
|
4.639
|
|
Ppm1h
|
protein phosphatase 1H (PP2C domain containing)
|
|
chr1_-_164408133
|
4.617
|
NM_001038619
NM_172646
|
Dnm3
|
dynamin 3
|
|
chr2_+_32576591
|
4.593
|
NM_013781
|
Sh2d3c
|
SH2 domain containing 3C
|
|
chr6_+_38868942
|
4.586
|
NM_011539
|
Tbxas1
|
thromboxane A synthase 1, platelet
|
|
chr7_-_56892061
|
4.575
|
NM_001005232
|
Dbx1
|
developing brain homeobox 1
|
|
chr1_-_164408123
|
4.566
|
|
Dnm3
|
dynamin 3
|
|
chr17_+_35399970
|
4.563
|
|
H2-D1
|
histocompatibility 2, D region locus 1
|
|
chr17_+_35400038
|
4.537
|
NM_010380
|
H2-D1
|
histocompatibility 2, D region locus 1
|
|
chr17_+_35607037
|
4.522
|
|
H2-Q10
|
histocompatibility 2, Q region locus 10
|
|
chr4_-_60594959
|
4.517
|
NM_001122647
|
Mup10
LOC100048884
Mup1
|
major urinary protein 10
novel member of the major urinary protein (Mup) gene family
major urinary protein 1
|
|
chr7_-_144506091
|
4.507
|
|
Ebf3
|
early B-cell factor 3
|
|
chr7_-_111507159
|
4.484
|
NM_175648
|
Trim30b
|
tripartite motif-containing 30B
|
|
chr19_-_21467250
|
4.479
|
|
Gda
|
guanine deaminase
|
|
chr3_+_52153283
|
4.475
|
|
Foxo1
|
forkhead box O1
|
|
chr7_-_148086341
|
4.463
|
NM_007751
|
Cox8b
|
cytochrome c oxidase, subunit VIIIb
|
|
chr6_+_88674697
|
4.462
|
NM_001166250
NM_001166251
|
Mgll
|
monoglyceride lipase
|
|
chr1_-_182175890
|
4.454
|
NM_011183
|
Psen2
|
presenilin 2
|
|
chr6_-_136889984
|
4.425
|
|
Arhgdib
|
Rho, GDP dissociation inhibitor (GDI) beta
|
|
chr2_-_155999858
|
4.397
|
|
Rbm39
|
RNA binding motif protein 39
|
|
chr18_+_63868337
|
4.395
|
NM_001014981
|
Wdr7
|
WD repeat domain 7
|
|
chr1_-_164408008
|
4.388
|
|
Dnm3
|
dynamin 3
|
|
chr7_+_107621474
|
4.382
|
NM_009464
|
Ucp3
|
uncoupling protein 3 (mitochondrial, proton carrier)
|
|
chr14_+_28051224
|
4.339
|
NM_027871
|
Arhgef3
|
Rho guanine nucleotide exchange factor (GEF) 3
|
|
chr2_-_126444378
|
4.335
|
NM_008230
|
Hdc
|
histidine decarboxylase
|