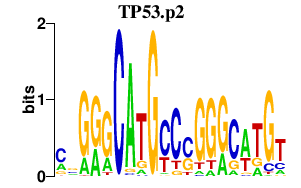
|
chr4_+_148478321
|
10.175
|
|
Dffa
|
DNA fragmentation factor, alpha subunit
|
|
chr4_+_148478238
|
9.257
|
NM_001025296
NM_010044
|
Dffa
|
DNA fragmentation factor, alpha subunit
|
|
chr9_-_123760992
|
9.173
|
NM_001110253
|
Fyco1
|
FYVE and coiled-coil domain containing 1
|
|
chr9_-_123760686
|
7.035
|
NM_148925
|
Fyco1
|
FYVE and coiled-coil domain containing 1
|
|
chr11_-_106163120
|
4.781
|
NM_008117
|
Gh
|
growth hormone
|
|
chr4_+_148478290
|
4.779
|
|
Dffa
|
DNA fragmentation factor, alpha subunit
|
|
chr1_-_21952022
|
4.128
|
NM_001160139
NM_023872
|
Kcnq5
|
potassium voltage-gated channel, subfamily Q, member 5
|
|
chr8_-_86265035
|
4.033
|
NM_001163029
NM_001163030
NM_001163031
NM_011925
|
Cd97
|
CD97 antigen
|
|
chr6_-_115708978
|
3.830
|
|
Tmem40
|
transmembrane protein 40
|
|
chr11_-_40568710
|
3.795
|
NM_009831
|
Ccng1
|
cyclin G1
|
|
chr9_-_34983552
|
3.284
|
NM_027030
|
Dcps
|
decapping enzyme, scavenger
|
|
chr7_-_148068161
|
2.889
|
NM_022433
|
Sirt3
|
sirtuin 3 (silent mating type information regulation 2, homolog) 3 (S. cerevisiae)
|
|
chr7_+_109416314
|
2.700
|
NM_009287
|
Stim1
|
stromal interaction molecule 1
|
|
chr6_-_115709004
|
2.616
|
NM_144805
|
Tmem40
|
transmembrane protein 40
|
|
chr1_-_17087821
|
2.612
|
NM_020604
|
Jph1
|
junctophilin 1
|
|
chr1_-_17087915
|
2.571
|
|
Jph1
|
junctophilin 1
|
|
chr11_+_60351159
|
2.555
|
NM_172943
|
Alkbh5
|
alkB, alkylation repair homolog 5 (E. coli)
|
|
chr1_-_21951497
|
2.547
|
|
Kcnq5
|
potassium voltage-gated channel, subfamily Q, member 5
|
|
chr12_+_77471173
|
2.465
|
NM_178744
|
Zbtb1
|
zinc finger and BTB domain containing 1
|
|
chr16_-_4523061
|
2.367
|
|
Srl
|
sarcalumenin
|
|
chr11_-_78959855
|
2.297
|
NM_013571
|
Ksr1
|
kinase suppressor of ras 1
|
|
chr11_-_78959969
|
2.293
|
|
Ksr1
|
kinase suppressor of ras 1
|
|
chr4_+_124419115
|
2.284
|
NM_008385
|
Inpp5b
|
inositol polyphosphate-5-phosphatase B
|
|
chr6_-_136610399
|
2.237
|
NM_025806
|
Plbd1
|
phospholipase B domain containing 1
|
|
chr12_+_113858217
|
2.235
|
NM_007421
|
Adssl1
|
adenylosuccinate synthetase like 1
|
|
chr16_-_10542689
|
2.224
|
|
Dexi
|
dexamethasone-induced transcript
|
|
chr11_-_40568524
|
2.040
|
|
Ccng1
|
cyclin G1
|
|
chr14_-_73725546
|
2.036
|
|
Rb1
|
retinoblastoma 1
|
|
chr14_-_73725576
|
2.032
|
NM_009029
|
Rb1
|
retinoblastoma 1
|
|
chr6_-_136610311
|
1.977
|
|
Plbd1
|
phospholipase B domain containing 1
|
|
chr11_-_109334909
|
1.962
|
NM_001029842
|
Slc16a6
|
solute carrier family 16 (monocarboxylic acid transporters), member 6
|
|
chr2_-_154900160
|
1.913
|
|
Ahcy
|
S-adenosylhomocysteine hydrolase
|
|
chr12_+_107248419
|
1.854
|
NM_001029843
NM_001029844
NM_011705
|
Vrk1
|
vaccinia related kinase 1
|
|
chr16_-_10543121
|
1.818
|
NM_021428
|
Dexi
|
dexamethasone-induced transcript
|
|
chr2_-_154900188
|
1.762
|
NM_016661
|
Ahcy
|
S-adenosylhomocysteine hydrolase
|
|
chr2_-_25877230
|
1.757
|
NM_133835
|
Ubac1
|
ubiquitin associated domain containing 1
|
|
chr4_-_106850812
|
1.649
|
NM_025617
|
2210012G02Rik
|
RIKEN cDNA 2210012G02 gene
|
|
chr9_-_121825422
|
1.641
|
NM_010012
|
Cyp8b1
|
cytochrome P450, family 8, subfamily b, polypeptide 1
|
|
chr12_+_107248496
|
1.628
|
|
Vrk1
|
vaccinia related kinase 1
|
|
chr2_-_154900132
|
1.597
|
|
Ahcy
|
S-adenosylhomocysteine hydrolase
|
|
chr17_-_31992708
|
1.582
|
NM_010831
|
Sik1
|
salt inducible kinase 1
|
|
chrX_-_35988248
|
1.575
|
|
C1galt1c1
|
C1GALT1-specific chaperone 1
|
|
chr12_-_105094033
|
1.549
|
|
Serpina1a
|
serine (or cysteine) peptidase inhibitor, clade A, member 1A
|
|
chr9_-_121825407
|
1.538
|
|
Cyp8b1
|
cytochrome P450, family 8, subfamily b, polypeptide 1
|
|
chr17_+_3326513
|
1.519
|
NM_001122998
|
Tiam2
|
T-cell lymphoma invasion and metastasis 2
|
|
chr1_+_54307526
|
1.495
|
NM_030025
|
Ccdc150
|
coiled-coil domain containing 150
|
|
chr19_-_47212695
|
1.482
|
NM_133746
|
Calhm2
|
calcium homeostasis modulator 2
|
|
chr11_-_5442157
|
1.391
|
NM_134033
|
Ccdc117
|
coiled-coil domain containing 117
|
|
chr11_-_12364754
|
1.381
|
|
Cobl
|
cordon-bleu
|
|
chr7_+_107855164
|
1.366
|
|
Fam168a
|
family with sequence similarity 168, member A
|
|
chr2_-_163223660
|
1.359
|
NM_021566
|
Jph2
|
junctophilin 2
|
|
chr8_+_97876182
|
1.344
|
NM_008609
|
Mmp15
|
matrix metallopeptidase 15
|
|
chr3_+_79395257
|
1.334
|
|
Ppid
|
peptidylprolyl isomerase D (cyclophilin D)
|
|
chr3_+_79395280
|
1.314
|
NM_026352
|
Ppid
|
peptidylprolyl isomerase D (cyclophilin D)
|
|
chr13_-_61037960
|
1.309
|
NM_007796
|
Ctla2a
|
cytotoxic T lymphocyte-associated protein 2 alpha
|
|
chr9_+_72833379
|
1.245
|
|
Ccpg1
|
cell cycle progression 1
|
|
chr15_-_77585658
|
1.240
|
NM_001081970
|
Apol8
|
apolipoprotein L 8
|
|
chr6_+_142705178
|
1.195
|
NM_009908
|
Cmas
|
cytidine monophospho-N-acetylneuraminic acid synthetase
|
|
chr17_+_36131397
|
1.182
|
|
A930015D03Rik
|
RIKEN cDNA A930015D03 gene
|
|
chr18_+_66618186
|
1.177
|
NM_021451
|
Pmaip1
|
phorbol-12-myristate-13-acetate-induced protein 1
|
|
chr8_+_75095914
|
1.170
|
NM_144534
|
Nwd1
Tmem38a
|
NACHT and WD repeat domain containing 1
transmembrane protein 38A
|
|
chr2_+_118489457
|
1.168
|
|
Pak6
|
p21 protein (Cdc42/Rac)-activated kinase 6
|
|
chr7_+_16894925
|
1.155
|
NM_133234
|
Bbc3
|
BCL2 binding component 3
|
|
chr4_+_155336386
|
1.129
|
NM_026125
|
Fam132a
|
family with sequence similarity 132, member A
|
|
chr8_+_96910392
|
1.090
|
NM_022331
|
Herpud1
|
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1
|
|
chr5_-_108416027
|
1.080
|
NM_026062
|
Fam69a
|
family with sequence similarity 69, member A
|
|
chr4_-_43512476
|
1.055
|
NM_001013377
|
E130306D19Rik
|
RIKEN cDNA E130306D19 gene
|
|
chr18_-_20160536
|
1.052
|
NM_007882
|
Dsc3
|
desmocollin 3
|
|
chr7_+_17428258
|
1.052
|
|
Prkd2
|
protein kinase D2
|
|
chr4_+_106851234
|
1.044
|
NM_029565
|
Tmem59
|
transmembrane protein 59
|
|
chr17_+_87682206
|
1.039
|
NM_028639
|
Ttc7
|
tetratricopeptide repeat domain 7
|
|
chr11_-_53520771
|
1.029
|
|
Rad50
|
RAD50 homolog (S. cerevisiae)
|
|
chr2_-_104334610
|
1.027
|
NM_001145824
NM_010434
|
Hipk3
|
homeodomain interacting protein kinase 3
|
|
chr9_+_72833327
|
1.026
|
NM_001114328
NM_028181
|
Ccpg1
|
cell cycle progression 1
|
|
chr12_-_71203819
|
1.009
|
NM_001081453
|
Nin
|
ninein
|
|
chr15_+_80808238
|
1.004
|
|
Sgsm3
|
small G protein signaling modulator 3
|
|
chr2_-_91864276
|
0.988
|
NM_011957
|
Creb3l1
|
cAMP responsive element binding protein 3-like 1
|
|
chr18_+_57514370
|
0.971
|
NM_028447
|
Prrc1
|
proline-rich coiled-coil 1
|
|
chr7_-_26022753
|
0.969
|
NM_001031667
|
Gsk3a
|
glycogen synthase kinase 3 alpha
|
|
chr9_+_51573434
|
0.967
|
NM_175535
|
Arhgap20
|
Rho GTPase activating protein 20
|
|
chr6_-_116411041
|
0.966
|
NM_009662
|
Alox5
|
arachidonate 5-lipoxygenase
|
|
chr12_+_3365295
|
0.946
|
|
Kif3c
|
kinesin family member 3C
|
|
chr6_+_66986637
|
0.945
|
|
E230016M11Rik
|
RIKEN cDNA E230016M11 gene
|
|
chr7_-_35437859
|
0.943
|
NM_146188
|
Kctd15
|
potassium channel tetramerisation domain containing 15
|
|
chr8_-_118230828
|
0.916
|
|
Maf
|
avian musculoaponeurotic fibrosarcoma (v-maf) AS42 oncogene homolog
|
|
chr12_+_84861415
|
0.909
|
NM_030246
|
Dcaf4
|
DDB1 and CUL4 associated factor 4
|
|
chr11_-_116099404
|
0.905
|
NM_025276
|
Evpl
|
envoplakin
|
|
chr15_+_80808194
|
0.905
|
NM_134091
|
Sgsm3
|
small G protein signaling modulator 3
|
|
chr3_-_135354500
|
0.884
|
NM_008689
|
Nfkb1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 1, p105
|
|
chr4_+_155336456
|
0.870
|
|
Fam132a
|
family with sequence similarity 132, member A
|
|
chr2_+_25098448
|
0.866
|
NM_176834
|
Rnf208
|
ring finger protein 208
|
|
chr2_-_180655611
|
0.863
|
NM_173761
|
Ythdf1
|
YTH domain family 1
|
|
chr2_-_25877064
|
0.860
|
|
Ubac1
|
ubiquitin associated domain containing 1
|
|
chr2_-_104334599
|
0.849
|
|
|
|
|
chr10_-_34138271
|
0.848
|
NM_176968
|
Nt5dc1
|
5'-nucleotidase domain containing 1
|
|
chr10_+_110601745
|
0.842
|
NM_001003717
NM_175489
|
Osbpl8
|
oxysterol binding protein-like 8
|
|
chr11_-_59104252
|
0.842
|
NM_009522
|
Wnt3a
|
wingless-related MMTV integration site 3A
|
|
chr4_+_62069853
|
0.842
|
|
Prpf4
|
PRP4 pre-mRNA processing factor 4 homolog (yeast)
|
|
chr2_+_91096984
|
0.823
|
|
Pacsin3
|
protein kinase C and casein kinase substrate in neurons 3
|
|
chr7_+_107855215
|
0.818
|
NM_178764
|
Fam168a
|
family with sequence similarity 168, member A
|
|
chr10_-_128002987
|
0.789
|
NM_011119
|
Pa2g4
|
proliferation-associated 2G4
|
|
chr8_+_3493117
|
0.785
|
NM_080461
|
Zfp358
|
zinc finger protein 358
|
|
chr4_-_57156022
|
0.781
|
NM_019427
|
Epb4.1l4b
|
erythrocyte protein band 4.1-like 4b
|
|
chr15_-_37890703
|
0.781
|
NM_199476
|
Rrm2b
|
ribonucleotide reductase M2 B (TP53 inducible)
|
|
chr14_-_55613384
|
0.777
|
NM_080728
|
Myh7
|
myosin, heavy polypeptide 7, cardiac muscle, beta
|
|
chr13_+_44826121
|
0.751
|
NM_021878
|
Jarid2
|
jumonji, AT rich interactive domain 2
|
|
chr1_+_132613903
|
0.746
|
NM_178691
|
Yod1
|
YOD1 OTU deubiquitinating enzyme 1 homologue (S. cerevisiae)
|
|
chr3_-_135354151
|
0.736
|
|
Nfkb1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 1, p105
|
|
chr17_-_33897428
|
0.732
|
NM_008997
|
Rab11b
|
RAB11B, member RAS oncogene family
|
|
chr15_-_78136047
|
0.730
|
|
Csf2rb2
|
colony stimulating factor 2 receptor, beta 2, low-affinity (granulocyte-macrophage)
|
|
chr18_+_4634926
|
0.713
|
NM_001081963
|
9430020K01Rik
|
RIKEN cDNA 9430020K01 gene
|
|
chr12_+_3365147
|
0.702
|
|
Kif3c
|
kinesin family member 3C
|
|
chr6_-_99616745
|
0.700
|
NM_025829
|
Eif4e3
|
eukaryotic translation initiation factor 4E member 3
|
|
chr9_+_65193874
|
0.697
|
NM_016688
|
Pdcd7
|
programmed cell death 7
|
|
chr4_+_62069801
|
0.685
|
NM_027297
|
Prpf4
|
PRP4 pre-mRNA processing factor 4 homolog (yeast)
|
|
chr7_+_134388553
|
0.685
|
|
Zfp771
|
zinc finger protein 771
|
|
chr11_-_63735652
|
0.668
|
NM_018805
|
Hs3st3b1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 3B1
|
|
chr4_-_118846799
|
0.658
|
NM_001145936
|
Zfp691
|
zinc finger protein 691
|
|
chr10_-_128002991
|
0.650
|
|
Pa2g4
|
proliferation-associated 2G4
|
|
chr17_+_74927634
|
0.644
|
NM_007566
|
Birc6
|
baculoviral IAP repeat-containing 6
|
|
chr11_+_78075242
|
0.643
|
NM_001002004
|
2610507B11Rik
|
RIKEN cDNA 2610507B11 gene
|
|
chr7_-_145589409
|
0.643
|
NM_183289
|
Tcerg1l
|
transcription elongation regulator 1-like
|
|
chr19_+_6401701
|
0.636
|
|
Rasgrp2
|
RAS, guanyl releasing protein 2
|
|
chr7_+_140050907
|
0.629
|
NM_198017
|
Fam175b
|
family with sequence similarity 175, member B
|
|
chr7_+_86956134
|
0.629
|
|
Mesp2
|
mesoderm posterior 2
|
|
chr4_+_140977621
|
0.626
|
|
Hspb7
|
heat shock protein family, member 7 (cardiovascular)
|
|
chr10_+_121078708
|
0.618
|
NM_177335
|
D930020B18Rik
|
RIKEN cDNA D930020B18 gene
|
|
chr16_-_13730474
|
0.617
|
NM_011987
|
Pla2g10
|
phospholipase A2, group X
|
|
chr8_-_23763885
|
0.610
|
NM_011130
|
Polb
|
polymerase (DNA directed), beta
|
|
chr19_-_7008874
|
0.609
|
NM_008431
|
Kcnk4
|
potassium channel, subfamily K, member 4
|
|
chr15_-_78633301
|
0.599
|
NM_130859
|
Card10
|
caspase recruitment domain family, member 10
|
|
chr19_-_45734544
|
0.599
|
NM_013907
|
Fbxw4
|
F-box and WD-40 domain protein 4
|
|
chr4_-_57155985
|
0.592
|
|
Epb4.1l4b
|
erythrocyte protein band 4.1-like 4b
|
|
chr18_+_82644510
|
0.591
|
NM_001025245
NM_010777
|
Mbp
|
myelin basic protein
|
|
chr4_+_151460980
|
0.590
|
|
Plekhg5
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5
|
|
chr17_+_50648736
|
0.580
|
NM_013880
|
Plcl2
|
phospholipase C-like 2
|
|
chr4_+_154338216
|
0.570
|
NM_172990
|
Pank4
|
pantothenate kinase 4
|
|
chr17_+_29227908
|
0.569
|
NM_001111099
|
Cdkn1a
|
cyclin-dependent kinase inhibitor 1A (P21)
|
|
chr14_+_57021419
|
0.567
|
NM_001033043
|
Rnf17
|
ring finger protein 17
|
|
chr1_+_195128163
|
0.567
|
NM_008484
|
Lamb3
|
laminin, beta 3
|
|
chr12_+_10397567
|
0.563
|
NM_023697
|
Rdh14
|
retinol dehydrogenase 14 (all-trans and 9-cis)
|
|
chr15_+_84847570
|
0.558
|
NM_023478
|
Upk3a
|
uroplakin 3A
|
|
chr11_-_86807216
|
0.555
|
NM_001005341
|
Ypel2
|
yippee-like 2 (Drosophila)
|
|
chr3_+_32716447
|
0.553
|
NM_001013024
|
Usp13
|
ubiquitin specific peptidase 13 (isopeptidase T-3)
|
|
chr19_-_34953614
|
0.552
|
|
Pank1
|
pantothenate kinase 1
|
|
chr13_-_60998747
|
0.552
|
NM_001145801
NM_007797
|
Ctla2b
|
cytotoxic T lymphocyte-associated protein 2 beta
|
|
chr13_-_53472732
|
0.544
|
|
Sptlc1
|
serine palmitoyltransferase, long chain base subunit 1
|
|
chr11_-_80593259
|
0.538
|
NM_177390
|
Myo1d
|
myosin ID
|
|
chr9_-_66823199
|
0.536
|
NM_030717
|
Lactb
|
lactamase, beta
|
|
chrX_-_35988178
|
0.535
|
|
C1galt1c1
|
C1GALT1-specific chaperone 1
|
|
chr9_-_36604816
|
0.527
|
|
Ei24
|
etoposide induced 2.4 mRNA
|
|
chr4_-_118293009
|
0.525
|
NM_026789
|
Wdr65
|
WD repeat domain 65
|
|
chr7_+_17428413
|
0.524
|
NM_178900
|
Prkd2
|
protein kinase D2
|
|
chr3_+_40698182
|
0.522
|
NM_026622
|
3110057O12Rik
|
RIKEN cDNA 3110057O12 gene
|
|
chr17_+_25325344
|
0.518
|
NM_001197024
|
Unkl
|
unkempt-like (Drosophila)
|
|
chr7_-_20043842
|
0.515
|
NM_172279
|
Mark4
|
MAP/microtubule affinity-regulating kinase 4
|
|
chr5_+_108561892
|
0.508
|
NM_028481
|
Ccdc18
|
coiled-coil domain containing 18
|
|
chr14_+_34899165
|
0.504
|
NM_013473
|
Anxa8
|
annexin A8
|
|
chr9_-_36604652
|
0.499
|
NM_007915
|
Ei24
|
etoposide induced 2.4 mRNA
|
|
chr9_-_57493393
|
0.497
|
|
Csk
|
c-src tyrosine kinase
|
|
chr7_+_6124083
|
0.489
|
NM_001146024
NM_028316
|
Zfp444
|
zinc finger protein 444
|
|
chr7_+_51640108
|
0.487
|
|
Syt3
|
synaptotagmin III
|
|
chr13_-_35085906
|
0.474
|
|
Peci
|
peroxisomal delta3, delta2-enoyl-Coenzyme A isomerase
|
|
chr4_-_49533914
|
0.473
|
NM_178603
|
Mrpl50
|
mitochondrial ribosomal protein L50
|
|
chr9_-_121614076
|
0.470
|
NM_001164562
NM_178677
|
Sec22c
|
SEC22 vesicle trafficking protein homolog C (S. cerevisiae)
|
|
chr9_-_69989285
|
0.468
|
NM_029784
|
Fam81a
|
family with sequence similarity 81, member A
|
|
chr11_+_60351469
|
0.467
|
|
Alkbh5
|
alkB, alkylation repair homolog 5 (E. coli)
|
|
chr1_-_138157449
|
0.467
|
NM_001101516
|
Gpr25
|
G protein-coupled receptor 25
|
|
chr4_+_140977613
|
0.465
|
|
Hspb7
|
heat shock protein family, member 7 (cardiovascular)
|
|
chr9_-_22272737
|
0.457
|
NM_018739
|
rp9
|
retinitis pigmentosa 9 (human)
|
|
chr13_+_38436987
|
0.454
|
|
Bmp6
|
bone morphogenetic protein 6
|
|
chr2_-_170322575
|
0.450
|
NM_009996
|
Cyp24a1
|
cytochrome P450, family 24, subfamily a, polypeptide 1
|
|
chrX_-_35988279
|
0.448
|
NM_021550
|
C1galt1c1
|
C1GALT1-specific chaperone 1
|
|
chrX_+_71422597
|
0.446
|
NM_031379
|
Tktl1
|
transketolase-like 1
|
|
chr4_-_150379857
|
0.441
|
|
Per3
|
period homolog 3 (Drosophila)
|
|
chr2_+_72123858
|
0.440
|
|
B230120H23Rik
|
RIKEN cDNA B230120H23 gene
|
|
chr12_+_3365102
|
0.440
|
NM_008445
|
Kif3c
|
kinesin family member 3C
|
|
chr18_-_67884185
|
0.440
|
NM_001127177
NM_008977
|
Ptpn2
|
protein tyrosine phosphatase, non-receptor type 2
|
|
chr5_-_135164426
|
0.439
|
NM_010717
|
Limk1
|
LIM-domain containing, protein kinase
|
|
chr18_+_82644535
|
0.435
|
|
Mbp
|
myelin basic protein
|
|
chr9_+_100978450
|
0.433
|
NM_001100451
|
Msl2
|
male-specific lethal 2 homolog (Drosophila)
|
|
chr13_-_53472719
|
0.432
|
NM_009269
|
Sptlc1
|
serine palmitoyltransferase, long chain base subunit 1
|
|
chr6_-_142519807
|
0.427
|
NM_008428
|
Kcnj8
|
potassium inwardly-rectifying channel, subfamily J, member 8
|
|
chr3_+_28680248
|
0.420
|
|
Eif5a2
|
eukaryotic translation initiation factor 5A2
|
|
chr7_-_112885071
|
0.419
|
|
Rrp8
|
ribosomal RNA processing 8, methyltransferase, homolog (yeast)
|
|
chr10_+_93103715
|
0.411
|
NM_021320
|
Ntn4
|
netrin 4
|
|
chr4_-_62069611
|
0.411
|
NM_139291
|
Cdc26
|
cell division cycle 26
|
|
chr7_-_35174519
|
0.406
|
NM_025948
|
Lsm14a
|
LSM14 homolog A (SCD6, S. cerevisiae)
|
|
chr4_-_115612507
|
0.405
|
NM_001025567
NM_130865
|
Dmbx1
|
diencephalon/mesencephalon homeobox 1
|
|
chr5_-_30399962
|
0.402
|
NM_021288
|
Tyms
|
thymidylate synthase
|
|
chr8_-_109572384
|
0.397
|
NM_026513
|
Pdf
|
peptide deformylase (mitochondrial)
|
|
chr4_-_141279303
|
0.391
|
NM_001017966
|
Ddi2
|
DNA-damage inducible protein 2
|
|
chr6_+_90554873
|
0.386
|
NM_027868
|
Slc41a3
|
solute carrier family 41, member 3
|
|
chr17_+_56904233
|
0.378
|
|
Dus3l
|
dihydrouridine synthase 3-like (S. cerevisiae)
|
|
chr9_+_32503600
|
0.377
|
NM_001038642
NM_011808
|
Ets1
|
E26 avian leukemia oncogene 1, 5' domain
|
|
chr11_-_53520811
|
0.375
|
NM_009012
|
Rad50
|
RAD50 homolog (S. cerevisiae)
|
|
chr4_-_133801438
|
0.375
|
NM_024215
|
Zfp593
|
zinc finger protein 593
|
|
chr4_+_132567420
|
0.372
|
NM_146155
|
Ahdc1
|
AT hook, DNA binding motif, containing 1
|
|
chr4_-_153471168
|
0.368
|
NM_001126330
NM_001126331
|
Trp73
|
transformation related protein 73
|
|
chr14_-_21167291
|
0.361
|
NM_027475
|
Ecd
|
ecdysoneless homolog (Drosophila)
|