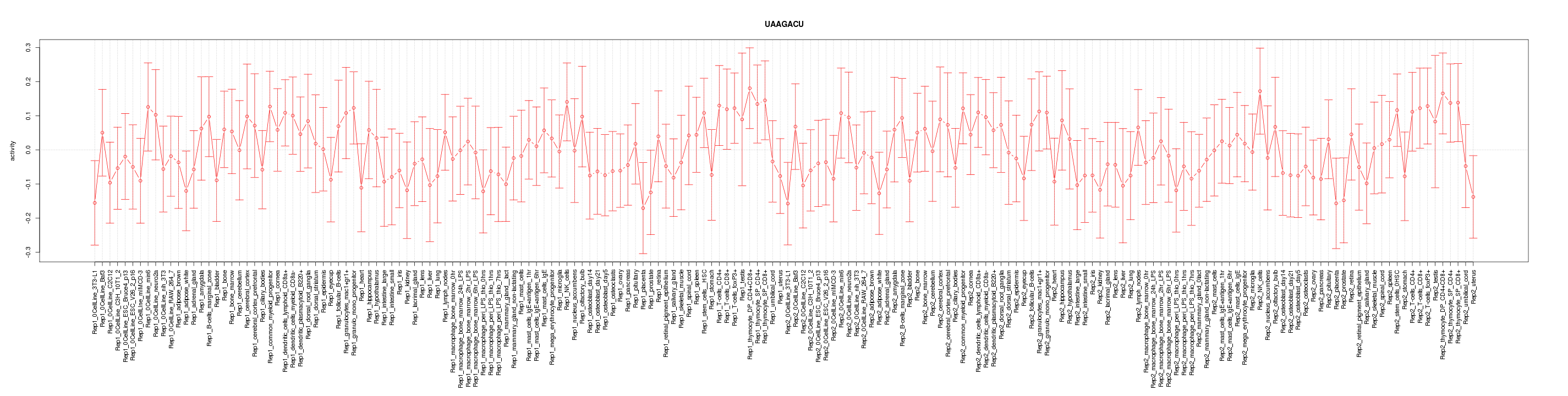
|
chr17_-_51951378
|
14.327
|
NM_001163630
|
Satb1
|
special AT-rich sequence binding protein 1
|
|
chr17_-_51965912
|
14.130
|
NM_001163631
|
Satb1
|
special AT-rich sequence binding protein 1
|
|
chr17_-_51972514
|
14.100
|
NM_001163632
|
Satb1
|
special AT-rich sequence binding protein 1
|
|
chr17_-_51971946
|
13.912
|
NM_009122
|
Satb1
|
special AT-rich sequence binding protein 1
|
|
chr16_+_45224449
|
9.804
|
NM_001037719
NM_177584
|
Btla
|
B and T lymphocyte associated
|
|
chr12_-_109241423
|
8.358
|
NM_001079883
NM_021399
|
Bcl11b
|
B-cell leukemia/lymphoma 11B
|
|
chr9_+_32503600
|
7.356
|
NM_001038642
NM_011808
|
Ets1
|
E26 avian leukemia oncogene 1, 5' domain
|
|
chr11_-_79317583
|
6.764
|
NM_019409
|
Omg
|
oligodendrocyte myelin glycoprotein
|
|
chr4_-_109959366
|
6.740
|
NM_001163399
NM_010488
|
Elavl4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr4_-_110024513
|
6.381
|
NM_001163397
|
Elavl4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr4_-_109960078
|
6.227
|
NM_001038698
|
Elavl4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr2_-_7002344
|
5.952
|
NM_001110229
NM_001110230
NM_001160292
|
Celf2
|
CUGBP, Elav-like family member 2
|
|
chr12_-_114067537
|
5.422
|
NM_028023
|
Cdca4
|
cell division cycle associated 4
|
|
chr10_+_89336253
|
4.823
|
NM_001128086
|
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B
|
|
chr9_+_72380046
|
4.460
|
NM_001033536
|
Rfx7
|
regulatory factor X, 7
|
|
chr4_-_86876328
|
4.395
|
NM_172426
|
Slc24a2
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 2
|
|
chr2_-_7317824
|
4.378
|
NM_001110231
|
Celf2
|
CUGBP, Elav-like family member 2
|
|
chr2_-_6805937
|
4.336
|
NM_001110228
NM_001160293
NM_010160
|
Celf2
|
CUGBP, Elav-like family member 2
|
|
chr11_-_78510874
|
4.284
|
NM_008702
|
Nlk
|
nemo like kinase
|
|
chr1_-_21952022
|
4.184
|
NM_001160139
NM_023872
|
Kcnq5
|
potassium voltage-gated channel, subfamily Q, member 5
|
|
chr6_-_99616745
|
3.985
|
NM_025829
|
Eif4e3
|
eukaryotic translation initiation factor 4E member 3
|
|
chr13_-_56236910
|
3.923
|
NM_001159513
NM_001159514
NM_001159515
NM_012015
|
H2afy
|
H2A histone family, member Y
|
|
chr2_-_7317513
|
3.851
|
NM_001110232
|
Celf2
|
CUGBP, Elav-like family member 2
|
|
chr16_-_78576898
|
3.757
|
NM_025967
|
D16Ertd472e
|
DNA segment, Chr 16, ERATO Doi 472, expressed
|
|
chr4_-_91066674
|
3.660
|
NM_207685
|
Elavl2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr11_-_86171026
|
3.646
|
NM_001080931
|
Med13
|
mediator complex subunit 13
|
|
chr6_+_77192615
|
3.599
|
NM_028880
|
Lrrtm1
|
leucine rich repeat transmembrane neuronal 1
|
|
chr4_-_91042997
|
3.589
|
NM_001177883
|
Elavl2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr4_-_133843139
|
3.455
|
NM_001163794
|
Pdik1l
|
PDLIM1 interacting kinase 1 like
|
|
chr4_-_91038729
|
3.357
|
NM_010486
NM_207686
|
Elavl2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr14_-_21365595
|
3.341
|
NM_008914
|
Ppp3cb
|
protein phosphatase 3, catalytic subunit, beta isoform
|
|
chr11_+_98769464
|
3.255
|
NM_011799
|
Cdc6
|
cell division cycle 6 homolog (S. cerevisiae)
|
|
chr4_-_86873866
|
3.242
|
NM_001110240
|
Slc24a2
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 2
|
|
chr5_-_9161670
|
3.225
|
NM_001110327
NM_011806
|
Dmtf1
|
cyclin D binding myb-like transcription factor 1
|
|
chr11_+_98769162
|
3.138
|
NM_001025779
|
Cdc6
|
cell division cycle 6 homolog (S. cerevisiae)
|
|
chr4_-_133843746
|
3.130
|
NM_146156
|
Pdik1l
|
PDLIM1 interacting kinase 1 like
|
|
chr10_+_90039435
|
3.091
|
NM_001177396
NM_001177398
NM_181398
|
Ipw
Anks1b
|
imprinted gene in the Prader-Willi syndrome region
ankyrin repeat and sterile alpha motif domain containing 1B
|
|
chr17_+_83750246
|
3.088
|
NM_001114361
NM_001114362
NM_199466
|
Eml4
|
echinoderm microtubule associated protein like 4
|
|
chr10_+_90038471
|
3.068
|
NM_001177397
|
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B
|
|
chr3_-_116126932
|
3.067
|
NM_001080818
NM_001173553
|
Cdc14a
|
CDC14 cell division cycle 14 homolog A (S. cerevisiae)
|
|
chr19_-_14672472
|
2.972
|
NM_011600
|
Tle4
|
transducin-like enhancer of split 4, homolog of Drosophila E(spl)
|
|
chr6_+_92041408
|
2.788
|
NM_011630
|
Nr2c2
|
nuclear receptor subfamily 2, group C, member 2
|
|
chr1_-_69732373
|
2.756
|
NM_011770
|
Ikzf2
|
IKAROS family zinc finger 2
|
|
chr15_-_84686558
|
2.743
|
NM_001081166
|
Phf21b
|
PHD finger protein 21B
|
|
chr13_+_109444370
|
2.653
|
NM_011056
|
Pde4d
|
phosphodiesterase 4D, cAMP specific
|
|
chr15_+_96117952
|
2.616
|
NM_175251
|
Arid2
|
AT rich interactive domain 2 (ARID, RFX-like)
|
|
chr3_+_94641479
|
2.613
|
NM_001165948
NM_172683
|
Pogz
|
pogo transposable element with ZNF domain
|
|
chr5_+_123794456
|
2.565
|
NM_029850
|
Bcl7a
|
B-cell CLL/lymphoma 7A
|
|
chr5_+_108494692
|
2.514
|
NM_013827
|
Mtf2
|
metal response element binding transcription factor 2
|
|
chr9_+_44925922
|
2.444
|
NM_001014761
|
Scn2b
|
sodium channel, voltage-gated, type II, beta
|
|
chr2_-_37502738
|
2.347
|
NM_009261
|
Strbp
|
spermatid perinuclear RNA binding protein
|
|
chr3_+_109143558
|
2.256
|
NM_020505
|
Vav3
|
vav 3 oncogene
|
|
chr14_+_79826199
|
2.249
|
NM_001024135
|
Kbtbd7
|
kelch repeat and BTB (POZ) domain containing 7
|
|
chr11_+_80290509
|
2.209
|
NM_009871
|
Cdk5r1
|
cyclin-dependent kinase 5, regulatory subunit 1 (p35)
|
|
chr5_-_23122181
|
2.194
|
NM_009274
|
Srpk2
|
serine/arginine-rich protein specific kinase 2
|
|
chr3_-_119486305
|
2.190
|
NM_019550
|
Ptbp2
|
polypyrimidine tract binding protein 2
|
|
chr9_+_86360760
|
2.139
|
NM_177208
|
Dopey1
|
dopey family member 1
|
|
chr18_+_86882439
|
2.135
|
NM_172633
|
Cbln2
|
cerebellin 2 precursor protein
|
|
chr1_+_160292470
|
2.127
|
NM_007495
|
Astn1
|
astrotactin 1
|
|
chr16_+_44173356
|
2.030
|
NM_001029889
|
Gm608
|
predicted gene 608
|
|
chr11_-_93747039
|
2.005
|
NM_001013375
|
Utp18
|
UTP18, small subunit (SSU) processome component, homolog (yeast)
|
|
chr12_+_33063653
|
1.997
|
NM_001162903
|
2010109K11Rik
|
RIKEN cDNA 2010109K11 gene
|
|
chr19_+_53966720
|
1.895
|
NM_011050
|
Pdcd4
|
programmed cell death 4
|
|
chr7_-_73834420
|
1.808
|
NM_080443
|
Asb7
|
ankyrin repeat and SOCS box-containing 7
|
|
chr3_+_109376824
|
1.805
|
NM_146139
|
Vav3
|
vav 3 oncogene
|
|
chr13_-_112280018
|
1.788
|
NM_001122963
NM_028487
|
Gpbp1
|
GC-rich promoter binding protein 1
|
|
chr12_+_50483869
|
1.787
|
NM_001160112
NM_008241
|
Foxg1
|
forkhead box G1
|
|
chr12_-_70782807
|
1.755
|
NM_001135559
|
Sos2
|
son of sevenless homolog 2 (Drosophila)
|
|
chr18_+_37127284
|
1.748
|
NM_007767
|
Pcdha6
|
protocadherin alpha 6
|
|
chr18_+_37303622
|
1.744
|
NM_001003672
|
Pcdhac2
|
protocadherin alpha subfamily C, 2
|
|
chr17_-_25152174
|
1.702
|
NM_020608
|
Cramp1l
|
Crm, cramped-like (Drosophila)
|
|
chr18_+_37089837
|
1.636
|
NM_054072
|
Pcdha1
|
protocadherin alpha 1
|
|
chr19_-_32786771
|
1.614
|
NM_026487
|
Atad1
|
ATPase family, AAA domain containing 1
|
|
chr18_+_37105758
|
1.607
|
NM_138662
|
Pcdha3
|
protocadherin alpha 3
|
|
chr8_+_119873069
|
1.602
|
NM_028941
|
4933407C03Rik
|
RIKEN cDNA 4933407C03 gene
|
|
chrX_-_69901382
|
1.593
|
NM_008067
|
Gabra3
|
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 3
|
|
chr11_-_106649652
|
1.592
|
NM_007840
|
Ddx5
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 5
|
|
chr18_+_37120093
|
1.585
|
NM_009959
|
Pcdha5
|
protocadherin alpha 5
|
|
chr18_-_81183260
|
1.499
|
NM_178280
|
Sall3
|
sal-like 3 (Drosophila)
|
|
chr8_+_107694536
|
1.493
|
NM_001161456
NM_001161457
NM_001161458
NM_022309
|
Cbfb
|
core binding factor beta
|
|
chr18_+_37152024
|
1.465
|
NM_201243
|
Pcdha8
|
protocadherin alpha 8
|
|
chr3_-_89767429
|
1.438
|
NM_001081182
|
Atp8b2
|
ATPase, class I, type 8B, member 2
|
|
chr18_+_37157533
|
1.430
|
NM_138661
|
Pcdha9
|
protocadherin alpha 9
|
|
chr17_+_88840048
|
1.421
|
NM_180974
|
Foxn2
|
forkhead box N2
|
|
chr18_+_37179802
|
1.416
|
NM_138663
|
Pcdha12
|
protocadherin alpha 12
|
|
chr8_+_41393330
|
1.372
|
NM_030110
|
Efha2
|
EF-hand domain family, member A2
|
|
chr8_+_93352723
|
1.369
|
NM_177224
|
Chd9
|
chromodomain helicase DNA binding protein 9
|
|
chrX_-_154481041
|
1.307
|
NM_177751
|
Cnksr2
|
connector enhancer of kinase suppressor of Ras 2
|
|
chr18_+_37098858
|
1.283
|
NM_198117
|
Pcdha2
|
protocadherin alpha 2
|
|
chr11_-_97436439
|
1.226
|
NM_018873
|
Srcin1
|
SRC kinase signaling inhibitor 1
|
|
chr12_-_92828896
|
1.208
|
NM_175335
|
Gtf2a1
|
general transcription factor II A, 1
|
|
chr2_+_152673699
|
1.201
|
NM_001141975
NM_001141976
NM_001141977
NM_001141978
NM_028109
|
Tpx2
|
TPX2, microtubule-associated protein homolog (Xenopus laevis)
|
|
chr18_+_83080270
|
1.152
|
NM_183033
|
Zfp516
|
zinc finger protein 516
|
|
chr18_+_37133455
|
1.086
|
NM_009957
|
Pcdha7
|
protocadherin alpha 7
|
|
chr11_-_107772661
|
1.063
|
NM_080644
|
Cacng5
|
calcium channel, voltage-dependent, gamma subunit 5
|
|
chr14_-_67487226
|
1.054
|
NM_009955
|
Dpysl2
|
dihydropyrimidinase-like 2
|
|
chr4_+_107474842
|
1.032
|
NM_001080926
|
Lrp8
|
low density lipoprotein receptor-related protein 8, apolipoprotein e receptor
|
|
chr4_-_133684599
|
1.009
|
NM_080559
|
Sh3bgrl3
|
SH3 domain binding glutamic acid-rich protein-like 3
|
|
chr7_-_128850659
|
0.967
|
NM_001033173
|
Usp31
|
ubiquitin specific peptidase 31
|
|
chr13_-_48720941
|
0.963
|
NM_207232
|
Ptpdc1
|
protein tyrosine phosphatase domain containing 1
|
|
chr18_+_37170365
|
0.931
|
NM_009960
|
Pcdha11
|
protocadherin alpha 11
|
|
chr8_+_119780918
|
0.887
|
NM_001163262
|
4933407C03Rik
|
RIKEN cDNA 4933407C03 gene
|
|
chr8_-_8690519
|
0.877
|
NM_176849
|
Arglu1
|
arginine and glutamate rich 1
|
|
chr18_+_32322742
|
0.875
|
NM_011946
|
Map3k2
|
mitogen-activated protein kinase kinase kinase 2
|
|
chr2_+_67955769
|
0.846
|
NM_020283
|
B3galt1
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1
|
|
chr17_-_6860516
|
0.805
|
NM_001166627
NM_001166630
|
Dynlt1f
Dynlt1c
Dynlt1b
|
dynein light chain Tctex-type 1F
dynein light chain Tctex-type 1C
dynein light chain Tctex-type 1D
|
|
chr17_+_6600795
|
0.796
|
NM_001166627
NM_001166630
|
Dynlt1f
Dynlt1c
Dynlt1b
|
dynein light chain Tctex-type 1F
dynein light chain Tctex-type 1C
dynein light chain Tctex-type 1D
|
|
chr15_-_93105514
|
0.786
|
NM_001033275
|
Gxylt1
|
glucoside xylosyltransferase 1
|
|
chr17_-_6654924
|
0.785
|
NM_001166627
NM_001166630
|
Dynlt1f
Dynlt1c
Dynlt1b
|
dynein light chain Tctex-type 1F
dynein light chain Tctex-type 1C
dynein light chain Tctex-type 1D
|
|
chr10_-_23069692
|
0.761
|
NM_010167
|
Eya4
|
eyes absent 4 homolog (Drosophila)
|
|
chr19_+_53977788
|
0.754
|
NM_001168491
NM_001168492
|
Pdcd4
|
programmed cell death 4
|
|
chr11_+_44332426
|
0.753
|
NM_028862
|
Rnf145
|
ring finger protein 145
|
|
chr4_+_140703825
|
0.738
|
NM_172518
|
Fbxo42
|
F-box protein 42
|
|
chr11_-_4494815
|
0.730
|
NM_028860
|
Mtmr3
|
myotubularin related protein 3
|
|
chr3_+_89324085
|
0.669
|
NM_080466
|
Kcnn3
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3
|
|
chr1_-_135557934
|
0.612
|
NM_144530
|
Zbed6
Zc3h11a
|
zinc finger, BED domain containing 6
zinc finger CCCH type containing 11A
|
|
chr4_+_105988790
|
0.602
|
NM_183225
|
Usp24
|
ubiquitin specific peptidase 24
|
|
chr8_-_119256732
|
0.555
|
NM_029441
|
Cdyl2
|
chromodomain protein, Y chromosome-like 2
|
|
chr9_+_21172314
|
0.536
|
NM_001042707
NM_001042708
NM_001042709
NM_010561
|
Ilf3
|
interleukin enhancer binding factor 3
|
|
chr11_+_54251680
|
0.491
|
NM_173753
|
Fnip1
|
folliculin interacting protein 1
|
|
chr16_+_33380860
|
0.483
|
NM_011749
|
Zfp148
|
zinc finger protein 148
|
|
chr4_+_137773045
|
0.483
|
NM_001122897
|
Hp1bp3
|
heterochromatin protein 1, binding protein 3
|
|
chr18_+_36838868
|
0.458
|
NM_001083317
NM_026404
|
Slc35a4
|
solute carrier family 35, member A4
|
|
chr11_-_82642528
|
0.443
|
NM_026097
|
Rffl
|
ring finger and FYVE like domain containing protein
|
|
chr11_+_97311458
|
0.432
|
NM_021493
|
Arhgap23
|
Rho GTPase activating protein 23
|
|
chr2_+_127951773
|
0.427
|
NM_009754
NM_207680
NM_207681
|
Bcl2l11
|
BCL2-like 11 (apoptosis facilitator)
|
|
chr9_+_86916846
|
0.406
|
NM_024195
|
Cyb5r4
|
cytochrome b5 reductase 4
|
|
chr9_-_70504934
|
0.381
|
NM_172772
|
Fam63b
|
family with sequence similarity 63, member B
|
|
chr7_+_56214442
|
0.379
|
NM_001111016
|
Nav2
|
neuron navigator 2
|
|
chr14_-_122196929
|
0.375
|
NM_134074
|
Dock9
|
dedicator of cytokinesis 9
|
|
chr8_-_97830469
|
0.366
|
NM_001195413
NM_145601
|
Cngb1
|
cyclic nucleotide gated channel beta 1
|
|
chr1_-_24594228
|
0.316
|
NM_007733
|
Col19a1
|
collagen, type XIX, alpha 1
|
|
chr14_-_32390502
|
0.288
|
NM_009937
|
Colq
|
collagen-like tail subunit (single strand of homotrimer) of asymmetric acetylcholinesterase
|
|
chr4_+_137772474
|
0.286
|
NM_001122896
NM_010470
|
Hp1bp3
|
heterochromatin protein 1, binding protein 3
|
|
chr3_-_89716963
|
0.258
|
NM_010559
|
Il6ra
|
interleukin 6 receptor, alpha
|
|
chr12_-_92828018
|
0.226
|
NM_031391
|
Gtf2a1
|
general transcription factor II A, 1
|
|
chr9_+_111174321
|
0.207
|
NM_175266
|
Epm2aip1
|
EPM2A (laforin) interacting protein 1
|
|
chr1_-_121733647
|
0.205
|
NM_019933
|
Ptpn4
|
protein tyrosine phosphatase, non-receptor type 4
|
|
chr11_-_66339627
|
0.200
|
NM_001034874
|
Shisa6
|
shisa homolog 6 (Xenopus laevis)
|
|
chr18_+_37249789
|
0.198
|
NM_001003671
|
Pcdhac1
|
protocadherin alpha subfamily C, 1
|
|
chr9_-_59373258
|
0.196
|
NM_178388
|
Tmem202
|
transmembrane protein 202
|
|
chr8_-_80960532
|
0.193
|
NM_138944
|
Pou4f2
|
POU domain, class 4, transcription factor 2
|
|
chr1_+_133724553
|
0.161
|
NM_173865
|
Slc41a1
|
solute carrier family 41, member 1
|
|
chr5_+_67999118
|
0.097
|
NM_001033415
|
Shisa3
|
shisa homolog 3 (Xenopus laevis)
|
|
chr17_+_29686744
|
0.062
|
NM_198647
|
Tbc1d22b
|
TBC1 domain family, member 22B
|
|
chr9_+_44187572
|
0.049
|
NM_021395
|
Hyou1
|
hypoxia up-regulated 1
|
|
chr4_+_116393559
|
0.028
|
NM_146151
|
Tesk2
|
testis-specific kinase 2
|
|
chr14_-_122138082
|
0.027
|
NM_001128307
NM_001128308
|
Dock9
|
dedicator of cytokinesis 9
|
|
chr16_-_15594609
|
0.021
|
NM_001159351
NM_023585
|
Ube2v2
|
ubiquitin-conjugating enzyme E2 variant 2
|
|
chr1_-_51535232
|
0.019
|
NM_028696
|
Obfc2a
|
oligonucleotide/oligosaccharide-binding fold containing 2A
|