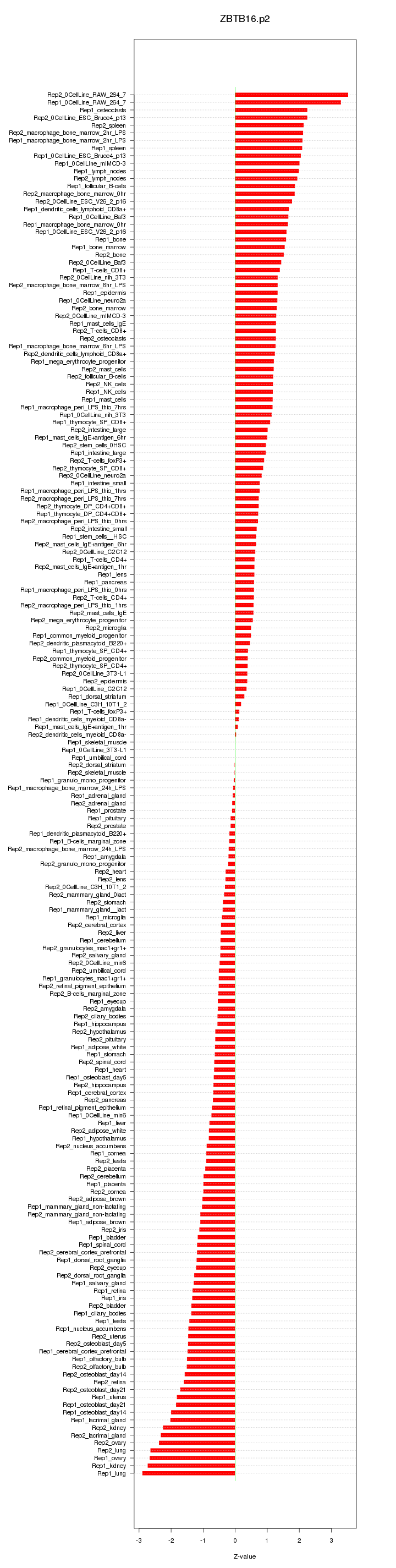
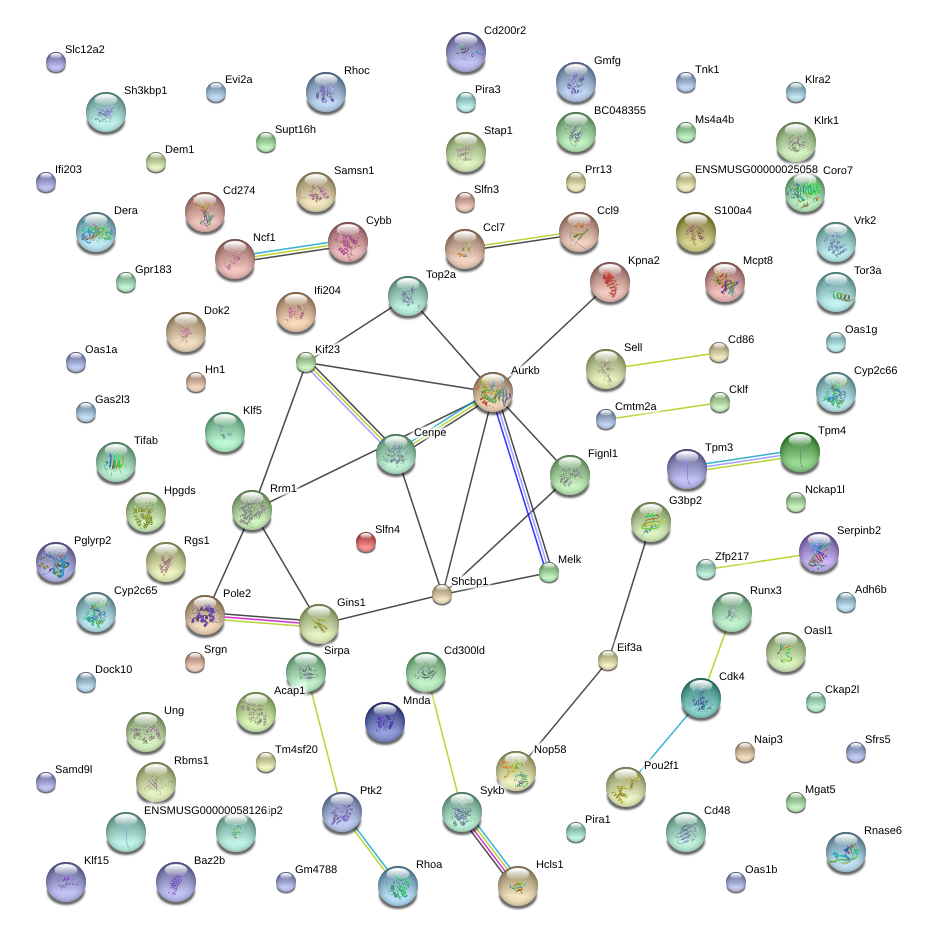
|
chr5_+_115373246
|
27.392
|
NM_145209
|
Oasl1
|
2'-5' oligoadenylate synthetase-like 1
|
|
chr5_+_115373272
|
23.061
|
|
Oasl1
|
2'-5' oligoadenylate synthetase-like 1
|
|
chr4_+_44313865
|
19.964
|
|
Melk
|
maternal embryonic leucine zipper kinase
|
|
chr11_+_82988687
|
19.282
|
NM_011410
|
Slfn4
|
schlafen 4
|
|
chr4_+_44313764
|
18.613
|
NM_010790
|
Melk
|
maternal embryonic leucine zipper kinase
|
|
chr10_-_61960613
|
16.326
|
|
Srgn
|
serglycin
|
|
chr2_-_129094163
|
14.151
|
|
Ckap2l
|
cytoskeleton associated protein 2-like
|
|
chr1_+_109407998
|
13.631
|
NM_001174170
NM_011111
|
Serpinb2
|
serine (or cysteine) peptidase inhibitor, clade B, member 2
|
|
chr11_+_81859212
|
12.696
|
NM_013654
|
Ccl7
|
chemokine (C-C motif) ligand 7
|
|
chr11_+_68859125
|
12.281
|
NM_011496
|
Aurkb
|
aurora kinase B
|
|
chr1_-_175805812
|
11.992
|
|
Mndal
|
myeloid nuclear differentiation antigen like
|
|
chr8_-_124948837
|
11.969
|
|
|
|
|
chr19_+_29441927
|
11.882
|
NM_021893
|
Cd274
|
CD274 antigen
|
|
chr1_-_158585297
|
11.642
|
|
Tor3a
|
torsin family 3, member A
|
|
chr11_-_83388330
|
11.615
|
|
Ccl9
|
chemokine (C-C motif) ligand 9
|
|
chr19_+_11518042
|
11.352
|
NM_021718
|
Ms4a4b
|
membrane-spanning 4-domains, subfamily A, member 4B
|
|
chr12_+_82049290
|
11.206
|
|
Srsf5
|
serine/arginine-rich splicing factor 5
|
|
chr11_-_98862163
|
10.940
|
|
Top2a
|
topoisomerase (DNA) II alpha
|
|
chr15_+_103284200
|
10.793
|
NM_153505
|
Nckap1l
|
NCK associated protein 1 like
|
|
chr1_+_173612132
|
9.845
|
|
Cd48
|
CD48 antigen
|
|
chr5_+_115373333
|
9.789
|
|
Oasl1
|
2'-5' oligoadenylate synthetase-like 1
|
|
chr17_-_32557807
|
9.712
|
|
Pglyrp2
|
peptidoglycan recognition protein 2
|
|
chr1_+_173612173
|
9.664
|
NM_007649
|
Cd48
|
CD48 antigen
|
|
chr10_-_88875526
|
9.550
|
|
Gas2l3
|
growth arrest-specific 2 like 3
|
|
chr16_+_44811858
|
9.360
|
|
Cd200r4
|
CD200 receptor 4
|
|
chr10_+_126503214
|
9.221
|
|
Cdk4
|
cyclin-dependent kinase 4
|
|
chr7_+_4225744
|
9.168
|
NM_011093
NM_008848
|
Pira6
|
paired-Ig-like receptor A6
|
|
chr17_-_32557916
|
8.995
|
NM_021319
|
Pglyrp2
|
peptidoglycan recognition protein 2
|
|
chr15_+_102289600
|
8.959
|
NM_001170911
NM_025385
|
Prr13
|
proline rich 13
|
|
chr16_-_36666078
|
8.559
|
NM_019388
|
Cd86
|
CD86 antigen
|
|
chr16_-_75909440
|
8.444
|
NM_023380
|
Samsn1
|
SAM domain, SH3 domain and nuclear localization signals, 1
|
|
chr7_+_29222465
|
8.442
|
NM_022024
|
Gmfg
|
glia maturation factor, gamma
|
|
chr6_+_137703029
|
8.199
|
NM_172733
|
Dera
|
2-deoxyribose-5-phosphate aldolase homolog (C. elegans)
|
|
chr5_+_34014563
|
8.192
|
|
|
|
|
chr14_-_122364309
|
8.168
|
NM_183031
|
Gpr183
|
G protein-coupled receptor 183
|
|
chr6_-_131197295
|
8.151
|
NM_001170851
NM_008462
|
Klra2
|
killer cell lectin-like receptor, subfamily A, member 2
|
|
chr5_+_86500770
|
8.091
|
NM_019992
|
Stap1
|
signal transducing adaptor family member 1
|
|
chr13_+_52678805
|
8.029
|
NM_001198977
|
Sykb
|
spleen tyrosine kinase
|
|
chr2_+_129442079
|
7.867
|
|
Sirpa
|
signal-regulatory protein alpha
|
|
chr6_-_3295980
|
7.859
|
|
9530028C05
|
hypothetical protein 9530028C05
|
|
chr8_+_74662546
|
7.813
|
|
Tpm4
|
tropomyosin 4
|
|
chr6_-_129572590
|
7.766
|
NM_033078
|
Klrk1
|
killer cell lectin-like receptor subfamily K, member 1
|
|
chr1_-_146096156
|
7.763
|
NM_015811
|
Rgs1
|
regulator of G-protein signaling 1
|
|
chr2_-_60589059
|
7.749
|
|
Rbms1
|
RNA binding motif, single stranded interacting protein 1
|
|
chr5_-_134705423
|
7.733
|
NM_010876
|
Ncf1
|
neutrophil cytosolic factor 1
|
|
chr4_+_128688217
|
7.481
|
|
|
|
|
chr8_-_4779533
|
7.258
|
NM_011369
|
Shcbp1
|
Shc SH2-domain binding protein 1
|
|
chr2_+_150735306
|
7.183
|
NM_001163476
NM_027014
|
Gins1
|
GINS complex subunit 1 (Psf1 homolog)
|
|
chr11_+_83037063
|
7.065
|
|
AI662270
|
expressed sequence AI662270
|
|
chrX_+_156278399
|
6.950
|
NM_001135728
|
Sh3kbp1
|
SH3-domain kinase binding protein 1
|
|
chr14_+_51748742
|
6.918
|
NM_030098
|
Rnase6
|
ribonuclease, RNase A family, 6
|
|
chr7_-_25873560
|
6.841
|
|
|
|
|
chr2_-_169956719
|
6.813
|
NM_001033299
|
Zfp217
|
zinc finger protein 217
|
|
chr16_-_4679664
|
6.782
|
NM_030205
|
Coro7
|
coronin 7
|
|
chr4_+_134676559
|
6.670
|
NM_019732
|
Runx3
|
runt related transcription factor 3
|
|
chr4_-_120597563
|
6.601
|
NM_001160043
NM_028457
|
Dem1
|
defects in morphology 1 homolog (S. cerevisiae)
|
|
chr3_+_90407688
|
6.549
|
NM_011311
|
S100a4
|
S100 calcium binding protein A4
|
|
chr11_-_69709003
|
6.471
|
NM_153788
|
Acap1
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 1
|
|
chr11_-_106860803
|
6.331
|
NM_010655
|
Kpna2
|
karyopherin (importin) alpha 2
|
|
chr9_+_108239210
|
6.308
|
|
Rhoa
|
ras homolog gene family, member A
|
|
chr1_+_59741800
|
6.242
|
NM_018868
|
Nop58
|
NOP58 ribonucleoprotein homolog (yeast)
|
|
chr14_+_71174110
|
6.235
|
NM_010071
|
Dok2
|
docking protein 2
|
|
chr3_+_89904553
|
6.132
|
|
Tpm3
|
tropomyosin 3, gamma
|
|
chr3_+_134875526
|
6.126
|
NM_173762
|
Cenpe
|
centromere protein E
|
|
chr6_-_129187965
|
6.082
|
|
2310001H17Rik
|
RIKEN cDNA 2310001H17 gene
|
|
chr1_+_165992203
|
6.017
|
NM_001164059
NM_011346
|
Sell
|
selectin, lymphocyte
|
|
chr14_-_52780511
|
5.923
|
|
Supt16h
|
suppressor of Ty 16 homolog (S. cerevisiae)
|
|
chr16_+_44867209
|
5.923
|
NM_206535
|
Cd200r2
|
Cd200 receptor 2
|
|
chr1_-_82765022
|
5.889
|
NM_025453
|
Tm4sf20
|
transmembrane 4 L six family member 20
|
|
chr16_+_36935029
|
5.872
|
NM_008225
|
Hcls1
|
hematopoietic cell specific Lyn substrate 1
|
|
chr17_+_46633703
|
5.769
|
NM_207161
|
BC048355
|
cDNA sequence BC048355
|
|
chrX_-_9014351
|
5.670
|
|
Cybb
|
cytochrome b-245, beta polypeptide
|
|
chr19_+_39135495
|
5.654
|
NM_028191
|
Cyp2c65
|
cytochrome P450, family 2, subfamily c, polypeptide 65
|
|
chr1_-_80661844
|
5.648
|
|
Dock10
|
dedicator of cytokinesis 10
|
|
chr11_-_115375672
|
5.573
|
NM_008258
|
Hn1
|
hematological and neurological expressed sequence 1
|
|
chr11_-_83391984
|
5.545
|
|
Ccl9
|
chemokine (C-C motif) ligand 9
|
|
chr14_-_56704033
|
5.507
|
NM_008572
|
Mcpt8
|
mast cell protease 8
|
|
chrX_-_9046362
|
5.501
|
NM_007807
|
Cybb
|
cytochrome b-245, beta polypeptide
|
|
chr13_-_100972022
|
5.481
|
NM_001126182
NM_010872
|
Naip2
|
NLR family, apoptosis inhibitory protein 2
|
|
chr13_-_56280161
|
5.421
|
NM_001168615
NM_145976
|
Tifab
|
TRAF-interacting protein with forkhead-associated domain, family member B
|
|
chr11_-_26493919
|
5.421
|
NM_027260
|
Vrk2
|
vaccinia related kinase 2
|
|
chr12_-_70329149
|
5.382
|
NM_011133
|
Pole2
|
polymerase (DNA directed), epsilon 2 (p59 subunit)
|
|
chr7_+_109614267
|
5.374
|
|
Rrm1
|
ribonucleotide reductase M1
|
|
chr1_-_141677744
|
5.370
|
NM_001029977
NM_001160303
NM_001160304
|
EG214403
|
predicted gene, EG214403
|
|
chr1_-_167865030
|
5.350
|
NM_198933
NM_198934
|
Pou2f1
|
POU domain, class 2, transcription factor 1
|
|
chrX_+_83115611
|
5.340
|
NM_001163539
|
5430427O19Rik
|
RIKEN cDNA 5430427O19 gene
|
|
chr19_-_60842585
|
5.323
|
|
Eif3a
|
eukaryotic translation initiation factor 3, subunit A
|
|
chr11_-_11708917
|
5.309
|
NM_001163359
NM_001163360
NM_021891
|
Fignl1
|
fidgetin-like 1
|
|
chr5_-_121357467
|
5.273
|
NM_145211
|
Oas1a
|
2'-5' oligoadenylate synthetase 1A
|
|
chr6_-_3349415
|
5.258
|
|
Samd9l
|
sterile alpha motif domain containing 9-like
|
|
chr3_+_89904315
|
5.250
|
|
|
|
|
chr2_-_59963796
|
5.248
|
NM_001001182
|
Baz2b
|
bromodomain adjacent to zinc finger domain, 2B
|
|
chr8_+_106774774
|
5.174
|
NM_029295
NM_001037841
|
Cklf
|
chemokine-like factor
|
|
chr2_-_59963586
|
5.164
|
|
Baz2b
|
bromodomain adjacent to zinc finger domain, 2B
|
|
chr5_-_92483744
|
5.160
|
|
G3bp2
|
GTPase activating protein (SH3 domain) binding protein 2
|
|
chr5_+_114580397
|
5.156
|
NM_001040691
|
Ung
|
uracil DNA glycosylase
|
|
chr11_-_114851199
|
5.100
|
NM_145437
|
Cd300ld
|
CD300 molecule-like family member d
|
|
chr6_-_65094862
|
5.088
|
|
Hpgds
|
hematopoietic prostaglandin D synthase
|
|
chr11_-_79343891
|
5.080
|
|
Evi2a
|
ecotropic viral integration site 2a
|
|
chr9_-_61794540
|
5.063
|
NM_024245
|
Kif23
|
kinesin family member 23
|
|
chr14_+_75576599
|
5.033
|
|
Lcp1
|
lymphocyte cytosolic protein 1
|
|
chr11_-_83392109
|
5.019
|
|
Ccl9
|
chemokine (C-C motif) ligand 9
|
|
chrX_-_6765768
|
4.959
|
NM_016691
|
Clcn5
|
chloride channel 5
|
|
chr6_-_65094691
|
4.928
|
NM_019455
|
Hpgds
|
hematopoietic prostaglandin D synthase
|
|
chr11_-_82853975
|
4.907
|
NM_181542
|
Slfn10-ps
|
schlafen 10, pseudogene
|
|
chr13_+_52678866
|
4.887
|
|
Sykb
|
spleen tyrosine kinase
|
|
chr12_-_27129497
|
4.866
|
|
Rsad2
|
radical S-adenosyl methionine domain containing 2
|
|
chr6_-_3349470
|
4.771
|
NM_010156
|
Samd9l
|
sterile alpha motif domain containing 9-like
|
|
chr13_-_21879086
|
4.739
|
NM_178184
|
Hist1h2an
|
histone cluster 1, H2an
|
|
chr10_+_82359714
|
4.735
|
|
Txnrd1
|
thioredoxin reductase 1
|
|
chr5_+_8798145
|
4.723
|
NM_011075
|
Abcb1b
|
ATP-binding cassette, sub-family B (MDR/TAP), member 1B
|
|
chr17_-_43013320
|
4.713
|
NM_009847
|
Cd2ap
|
CD2-associated protein
|
|
chr6_+_129543505
|
4.708
|
NM_010654
|
Klrd1
|
killer cell lectin-like receptor, subfamily D, member 1
|
|
chr13_+_51740600
|
4.689
|
NM_025415
|
Cks2
|
CDC28 protein kinase regulatory subunit 2
|
|
chr3_-_135271247
|
4.686
|
|
Nfkb1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 1, p105
|
|
chr17_-_36169573
|
4.683
|
NM_010398
|
H2-T23
|
histocompatibility 2, T region locus 23
|
|
chr6_-_70067577
|
4.675
|
|
Gm10883
|
predicted gene 10883
|
|
chr2_+_90517371
|
4.662
|
NM_021512
|
Nup160
|
nucleoporin 160
|
|
chr5_+_122821868
|
4.638
|
|
Gpn3
|
GPN-loop GTPase 3
|
|
chr13_-_77230519
|
4.624
|
|
Ankrd32
|
ankyrin repeat domain 32
|
|
chr10_+_51210768
|
4.594
|
NM_013532
|
Lilrb4
|
leukocyte immunoglobulin-like receptor, subfamily B, member 4
|
|
chr2_-_25483295
|
4.590
|
|
Fcna
|
ficolin A
|
|
chr1_+_175350728
|
4.572
|
NM_001013779
|
Aim2
|
absent in melanoma 2
|
|
chr3_-_135271190
|
4.491
|
|
Nfkb1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 1, p105
|
|
chr16_-_15966921
|
4.483
|
|
2310008H04Rik
|
RIKEN cDNA 2310008H04 gene
|
|
chr11_-_79344055
|
4.479
|
NM_001033711
NM_010161
NM_146023
|
Evi2a
Evi2b
|
ecotropic viral integration site 2a
ecotropic viral integration site 2b
|
|
chr10_+_42580317
|
4.449
|
NM_172938
|
Scml4
|
sex comb on midleg-like 4 (Drosophila)
|
|
chr14_-_66452720
|
4.427
|
NM_028039
|
Esco2
|
establishment of cohesion 1 homolog 2 (S. cerevisiae)
|
|
chr4_+_107040388
|
4.361
|
NM_028355
|
Tmem48
|
transmembrane protein 48
|
|
chr1_-_175872522
|
4.333
|
NM_008328
NM_001045481
|
Ifi203
|
interferon activated gene 203
|
|
chr17_-_36248574
|
4.316
|
|
H2-T23
C920025E04Rik
|
histocompatibility 2, T region locus 23
RIKEN cDNA C920025E04 gene
|
|
chr3_-_135271220
|
4.275
|
|
Nfkb1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 1, p105
|
|
chr7_-_29678773
|
4.271
|
|
Actn4
|
actinin alpha 4
|
|
chr6_-_38238095
|
4.240
|
|
Zc3hav1l
|
zinc finger CCCH-type, antiviral 1-like
|
|
chr11_+_62416376
|
4.235
|
|
2410006H16Rik
|
RIKEN cDNA 2410006H16 gene
|
|
chr11_-_17853850
|
4.188
|
NM_026576
|
Etaa1
|
Ewing's tumor-associated antigen 1
|
|
chr6_-_130015247
|
4.166
|
NM_001039118
|
Klra4
Klra33
Klra18
|
killer cell lectin-like receptor, subfamily A, member 4
killer cell lectin-like receptor subfamily A member 33
killer cell lectin-like receptor, subfamily A, member 18
|
|
chr9_+_123429291
|
4.133
|
|
Limd1
|
LIM domains containing 1
|
|
chr19_+_39188387
|
4.126
|
NM_001011707
|
Cyp2c66
|
cytochrome P450, family 2, subfamily c, polypeptide 66
|
|
chr6_-_130183339
|
4.124
|
NM_001110323
|
Klra7
|
killer cell lectin-like receptor, subfamily A, member 7
|
|
chr5_-_130478687
|
4.097
|
NM_010368
|
Gusb
|
glucuronidase, beta
|
|
chr17_-_35312945
|
4.096
|
NM_019467
|
Aif1
|
allograft inflammatory factor 1
|
|
chr6_-_122806020
|
4.092
|
|
C3ar1
|
complement component 3a receptor 1
|
|
chr7_+_54106270
|
4.075
|
|
Ldha
|
lactate dehydrogenase A
|
|
chr2_-_180976728
|
4.004
|
NM_183162
|
BC006779
|
cDNA sequence BC006779
|
|
chr6_-_69557773
|
4.000
|
|
|
|
|
chr10_-_61970481
|
3.972
|
NM_011157
|
Srgn
|
serglycin
|
|
chr6_-_122806042
|
3.965
|
|
C3ar1
|
complement component 3a receptor 1
|
|
chr13_+_20277293
|
3.948
|
NM_080288
|
Elmo1
|
engulfment and cell motility 1, ced-12 homolog (C. elegans)
|
|
chr12_+_75028976
|
3.933
|
|
Hif1a
|
hypoxia inducible factor 1, alpha subunit
|
|
chr8_+_67471046
|
3.929
|
NM_001163747
NM_028427
|
Tmem192
|
transmembrane protein 192
|
|
chrX_-_163768423
|
3.923
|
NM_133211
|
Tlr7
|
toll-like receptor 7
|
|
chr13_+_43880475
|
3.918
|
NM_009856
|
Cd83
|
CD83 antigen
|
|
chr9_+_108953672
|
3.878
|
|
Shisa5
|
shisa homolog 5 (Xenopus laevis)
|
|
chr8_+_54597026
|
3.843
|
NM_001005847
|
Aga
|
aspartylglucosaminidase
|
|
chr11_-_86171026
|
3.842
|
NM_001080931
|
Med13
|
mediator complex subunit 13
|
|
chr2_+_152673699
|
3.818
|
NM_001141975
NM_001141976
NM_001141977
NM_001141978
NM_028109
|
Tpx2
|
TPX2, microtubule-associated protein homolog (Xenopus laevis)
|
|
chr5_-_92777875
|
3.814
|
NM_021274
|
Cxcl10
|
chemokine (C-X-C motif) ligand 10
|
|
chr5_+_145888588
|
3.779
|
|
Arpc1b
|
actin related protein 2/3 complex, subunit 1B
|
|
chr6_+_59158860
|
3.776
|
NM_001081145
|
Tigd2
|
tigger transposable element derived 2
|
|
chr19_+_11566527
|
3.775
|
NM_001164202
|
Gm8369
|
predicted gene 8369
|
|
chr1_-_58558259
|
3.750
|
NM_001025378
|
Orc2
|
origin recognition complex, subunit 2
|
|
chr11_-_83392129
|
3.741
|
NM_011338
|
Ccl9
|
chemokine (C-C motif) ligand 9
|
|
chr6_+_136467315
|
3.722
|
NM_019426
|
Atf7ip
|
activating transcription factor 7 interacting protein
|
|
chr13_-_60998747
|
3.718
|
NM_001145801
NM_007797
|
Ctla2b
|
cytotoxic T lymphocyte-associated protein 2 beta
|
|
chr4_-_132352272
|
3.708
|
NM_001033308
|
BC013712
|
cDNA sequence BC013712
|
|
chr11_-_40546863
|
3.692
|
NM_013552
|
Hmmr
|
hyaluronan mediated motility receptor (RHAMM)
|
|
chr6_+_129130632
|
3.656
|
NM_053109
|
Clec2d
|
C-type lectin domain family 2, member d
|
|
chr7_-_128217980
|
3.629
|
NM_030691
|
Igsf6
|
immunoglobulin superfamily, member 6
|
|
chr1_+_61034757
|
3.627
|
NM_017480
|
Icos
|
inducible T-cell co-stimulator
|
|
chrX_-_7182450
|
3.597
|
NM_138603
|
Ccdc22
|
coiled-coil domain containing 22
|
|
chr5_-_100392845
|
3.586
|
|
Hnrnpd
|
heterogeneous nuclear ribonucleoprotein D
|
|
chr7_+_28954717
|
3.586
|
NM_007991
|
Fbl
|
fibrillarin
|
|
chr2_-_26883441
|
3.560
|
NM_001177627
NM_172659
|
Slc2a6
|
solute carrier family 2 (facilitated glucose transporter), member 6
|
|
chr11_-_79337178
|
3.557
|
|
Evi2b
|
ecotropic viral integration site 2b
|
|
chr2_-_169968174
|
3.549
|
NM_001159683
|
Zfp217
|
zinc finger protein 217
|
|
chr4_-_124359695
|
3.535
|
|
Utp11l
|
UTP11-like, U3 small nucleolar ribonucleoprotein, (yeast)
|
|
chr13_-_55201655
|
3.526
|
NM_011307
|
Uimc1
|
ubiquitin interaction motif containing 1
|
|
chr6_+_41488583
|
3.515
|
|
|
|
|
chr6_-_122806122
|
3.498
|
NM_009779
|
C3ar1
|
complement component 3a receptor 1
|
|
chr5_+_116010796
|
3.435
|
|
Rplp0
|
ribosomal protein, large, P0
|
|
chr16_+_59148654
|
3.428
|
NM_146998
|
Olfr195
|
olfactory receptor 195
|
|
chr3_-_19164956
|
3.425
|
NM_008802
|
Pde7a
|
phosphodiesterase 7A
|
|
chr9_-_44152208
|
3.400
|
|
Hmbs
|
hydroxymethylbilane synthase
|
|
chr7_+_28345494
|
3.394
|
NM_001122767
|
2310022A10Rik
|
RIKEN cDNA 2310022A10 gene
|
|
chr19_+_55255374
|
3.381
|
NM_009348
|
Tectb
|
tectorin beta
|
|
chr11_+_3558702
|
3.369
|
NM_198162
|
Morc2a
|
microrchidia 2A
|
|
chr3_+_95466898
|
3.351
|
|
|
|
|
chr9_+_108953692
|
3.343
|
|
Shisa5
|
shisa homolog 5 (Xenopus laevis)
|
|
chr3_-_95161071
|
3.341
|
NM_001163641
NM_001163642
NM_018877
|
Setdb1
|
SET domain, bifurcated 1
|
|
chr9_+_64809499
|
3.336
|
|
2010321M09Rik
|
RIKEN cDNA 2010321M09 gene
|
|
chr7_-_4014717
|
3.333
|
NM_001113474
NM_178611
|
Lair1
|
leukocyte-associated Ig-like receptor 1
|
|
chr2_+_125977876
|
3.313
|
NM_026981
|
Dtwd1
|
DTW domain containing 1
|
|
chr19_+_29218114
|
3.305
|
|
|
|
|
chr2_-_167515255
|
3.288
|
|
Tmem189
|
transmembrane protein 189
|
|
chr1_+_173770709
|
3.288
|
NM_013489
|
Cd84
|
CD84 antigen
|
|
chr9_+_44887251
|
3.281
|
NM_001005421
|
Amica1
|
adhesion molecule, interacts with CXADR antigen 1
|
|
chr6_-_69193672
|
3.281
|
|
|
|
|
chr4_-_142718290
|
3.257
|
|
Prdm2
|
PR domain containing 2, with ZNF domain
|
|
chr7_+_148586221
|
3.255
|
|
Taldo1
|
transaldolase 1
|