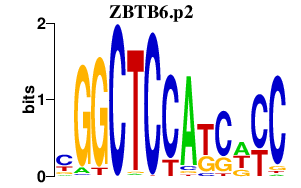
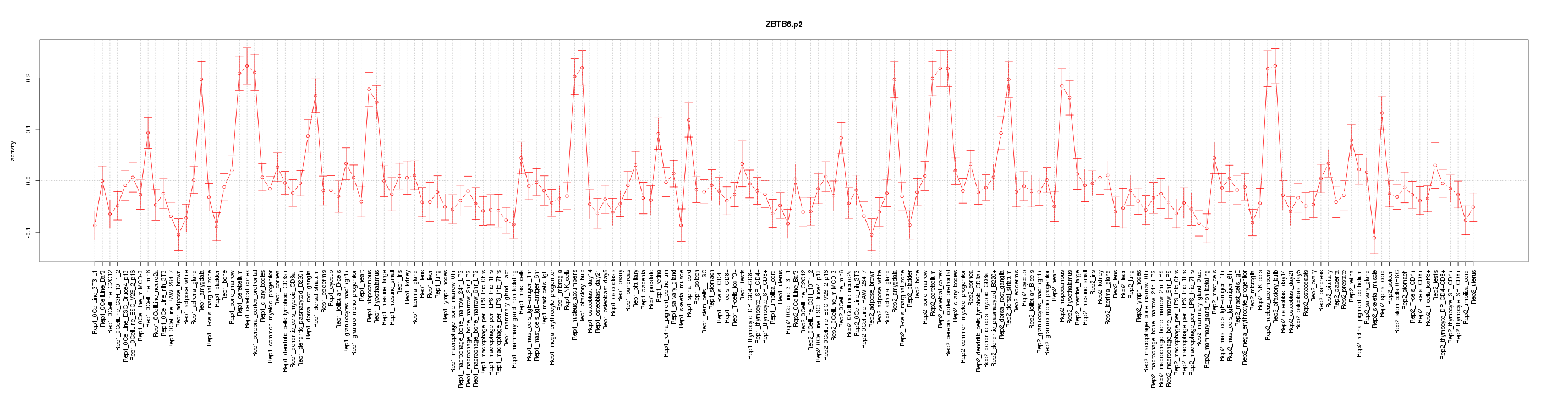
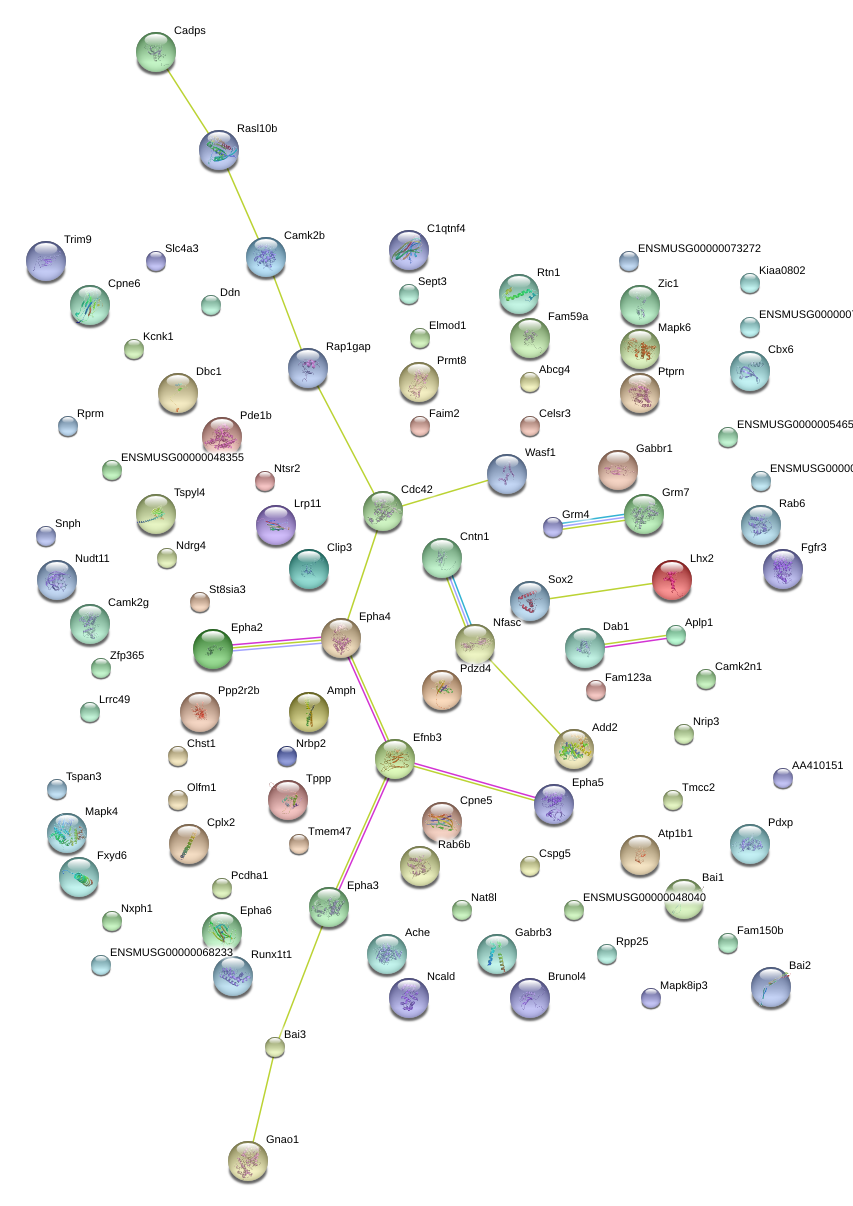
|
chr12_-_73509668
|
75.285
|
|
Rtn1
|
reticulon 1
|
|
chr12_-_73509537
|
59.232
|
|
Rtn1
|
reticulon 1
|
|
chr15_+_78744328
|
53.931
|
NM_020271
|
Pdxp
|
pyridoxal (pyridoxine, vitamin B6) phosphatase
|
|
chr18_-_25912171
|
42.968
|
|
Celf4
|
CUGBP, Elav-like family member 4
|
|
chr11_+_83223154
|
41.550
|
|
Rasl10b
|
RAS-like, family 10, member B
|
|
chr11_+_83223573
|
39.635
|
NM_001013386
|
Rasl10b
|
RAS-like, family 10, member B
|
|
chr3_+_34548916
|
35.741
|
NM_011443
|
Sox2
|
SRY-box containing gene 2
|
|
chr8_+_96334879
|
33.166
|
|
Gnao1
|
guanine nucleotide binding protein, alpha O
|
|
chr15_+_91881593
|
32.294
|
NM_001159648
|
Cntn1
|
contactin 1
|
|
chr12_-_73509925
|
30.816
|
NM_153457
|
Rtn1
|
reticulon 1
|
|
chr3_+_34549057
|
28.995
|
|
Sox2
|
SRY-box containing gene 2
|
|
chr6_+_110595586
|
28.955
|
NM_177328
|
Grm7
|
glutamate receptor, metabotropic 7
|
|
chr1_-_75260687
|
28.935
|
NM_008985
|
Ptprn
|
protein tyrosine phosphatase, receptor type, N
|
|
chr14_-_13655592
|
28.039
|
NM_001042617
NM_012061
|
Cadps
|
Ca2+-dependent secretion activator
|
|
chr14_-_13655558
|
27.740
|
|
Cadps
|
Ca2+-dependent secretion activator
|
|
chr15_-_98637560
|
27.186
|
|
Ddn
|
dendrin
|
|
chr7_-_31230564
|
26.384
|
|
Aplp1
|
amyloid beta (A4) precursor-like protein 1
|
|
chr18_-_43058544
|
25.164
|
NM_027531
|
Ppp2r2b
|
protein phosphatase 2 (formerly 2A), regulatory subunit B (PR 52), beta isoform
|
|
chr11_-_69373555
|
24.825
|
|
Efnb3
|
ephrin B3
|
|
chr7_-_31230577
|
24.817
|
NM_007467
|
Aplp1
|
amyloid beta (A4) precursor-like protein 1
|
|
chr18_+_37098858
|
23.951
|
NM_198117
|
Pcdha2
|
protocadherin alpha 2
|
|
chr10_+_34017221
|
23.713
|
NM_030203
|
Tspyl4
|
TSPY-like 4
|
|
chr11_-_69373707
|
23.551
|
NM_007911
|
Efnb3
|
ephrin B3
|
|
chr12_+_16660521
|
22.884
|
|
Ntsr2
|
neurotensin receptor 2
|
|
chr2_+_92439758
|
22.619
|
NM_023850
|
Chst1
|
carbohydrate (keratan sulfate Gal-6) sulfotransferase 1
|
|
chr1_-_166387998
|
22.421
|
|
Atp1b1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide
|
|
chr17_-_25073798
|
22.415
|
|
Mapk8ip3
|
mitogen-activated protein kinase 8 interacting protein 3
|
|
chr4_+_13670582
|
21.297
|
|
Runx1t1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr7_+_64845887
|
20.853
|
NM_001038701
|
Gabrb3
|
gamma-aminobutyric acid (GABA) A receptor, subunit beta 3
|
|
chr2_+_90729768
|
20.323
|
|
C1qtnf4
|
C1q and tumor necrosis factor related protein 4
|
|
chr13_+_19040215
|
20.316
|
NM_175007
|
Amph
|
amphiphysin
|
|
chr2_-_151458299
|
19.919
|
|
Snph
|
syntaphilin
|
|
chr14_+_56129440
|
19.876
|
|
Cpne6
|
copine VI
|
|
chr8_+_128519295
|
19.692
|
|
Kcnk1
|
potassium channel, subfamily K, member 1
|
|
chr7_+_107756292
|
19.586
|
|
Rab6
|
RAB6, member RAS oncogene family
|
|
chr1_-_134287712
|
19.260
|
NM_178874
|
Tmcc2
|
transmembrane and coiled-coil domains 2
|
|
chr17_-_27650285
|
19.221
|
|
Grm4
|
glutamate receptor, metabotropic 4
|
|
chrX_+_132567905
|
19.150
|
NM_029541
|
6530401D17Rik
|
RIKEN cDNA 6530401D17 gene
|
|
chr7_+_107756205
|
19.037
|
|
Rab6
|
RAB6, member RAS oncogene family
|
|
chr9_+_110148568
|
18.729
|
|
Cspg5
|
chondroitin sulfate proteoglycan 5
|
|
chr10_+_7309565
|
18.703
|
NM_172784
|
Lrp11
|
low density lipoprotein receptor-related protein 11
|
|
chr7_+_31076771
|
18.700
|
NM_001081114
|
Clip3
|
CAP-GLY domain containing linker protein 3
|
|
chr13_+_74146852
|
18.645
|
NM_182839
|
Tppp
|
tubulin polymerization promoting protein
|
|
chr14_+_60997104
|
18.549
|
NM_001164705
NM_028113
|
Fam123a
|
family with sequence similarity 123, member A
|
|
chr18_-_21458623
|
18.358
|
NM_001033445
|
Fam59a
|
family with sequence similarity 59, member A
|
|
chr2_-_151458256
|
18.084
|
NM_198214
|
Snph
|
syntaphilin
|
|
chr8_+_128519001
|
18.078
|
NM_008430
|
Kcnk1
|
potassium channel, subfamily K, member 1
|
|
chr10_+_40603330
|
18.059
|
NM_031877
|
Wasf1
|
WASP family 1
|
|
chr9_-_91259829
|
17.989
|
|
Zic1
|
zinc finger protein of the cerebellum 1
|
|
chr17_-_29374656
|
17.825
|
NM_153166
|
Cpne5
|
copine V
|
|
chr17_-_25073897
|
17.769
|
NM_001163447
NM_001163448
NM_001163449
NM_001163450
NM_001163451
NM_001163453
NM_013931
|
Mapk8ip3
|
mitogen-activated protein kinase 8 interacting protein 3
|
|
chr9_-_44096170
|
17.539
|
NM_138955
|
Abcg4
|
ATP-binding cassette, sub-family G (WHITE), member 4
|
|
chr4_-_68615382
|
17.471
|
NM_019967
|
Dbc1
|
deleted in bladder cancer 1 (human)
|
|
chr12_-_71448600
|
17.431
|
NM_001110202
NM_001110203
NM_053167
|
Trim9
|
tripartite motif-containing 9
|
|
chr15_+_82105322
|
17.421
|
NM_011889
|
Sept3
|
septin 3
|
|
chr7_+_31076852
|
17.280
|
|
Clip3
|
CAP-GLY domain containing linker protein 3
|
|
chr9_-_53822931
|
17.183
|
|
Elmod1
|
ELMO domain containing 1
|
|
chr4_-_136891931
|
17.009
|
|
Cdc42
|
cell division cycle 42 homolog (S. cerevisiae)
|
|
chr14_+_60997156
|
16.889
|
|
Fam123a
|
family with sequence similarity 123, member A
|
|
chr9_+_108728650
|
16.751
|
NM_080437
|
Celsr3
|
cadherin, EGF LAG seven-pass G-type receptor 3 (flamingo homolog, Drosophila)
|
|
chr10_-_67375332
|
16.724
|
NM_178679
|
Zfp365
|
zinc finger protein 365
|
|
chr17_+_37187550
|
16.325
|
|
Gabbr1
|
gamma-aminobutyric acid (GABA) B receptor, 1
|
|
chr1_-_53009546
|
16.204
|
NM_144953
|
1700019D03Rik
|
RIKEN cDNA 1700019D03 gene
|
|
chr15_-_75921924
|
16.045
|
|
Nrbp2
|
nuclear receptor binding protein 2
|
|
chr2_+_28061156
|
16.017
|
NM_001038612
NM_019498
|
Olfm1
|
olfactomedin 1
|
|
chrX_+_132528317
|
15.999
|
NM_029823
|
6530401D17Rik
2900062L11Rik
|
RIKEN cDNA 6530401D17 gene
RIKEN cDNA 2900062L11 gene
|
|
chr2_+_38206759
|
15.728
|
NM_010710
|
Lhx2
|
LIM homeobox protein 2
|
|
chr7_+_64846528
|
15.401
|
NM_008071
|
Gabrb3
|
gamma-aminobutyric acid (GABA) A receptor, subunit beta 3
|
|
chr2_-_53937785
|
15.368
|
NM_023396
|
Rprm
|
reprimo, TP53 dependent G2 arrest mediator candidate
|
|
chrX_+_78315963
|
15.309
|
NM_138751
|
Tmem47
|
transmembrane protein 47
|
|
chr1_+_75542840
|
15.048
|
NM_009208
|
Slc4a3
|
solute carrier family 4 (anion exchanger), member 3
|
|
chrX_+_78316003
|
14.982
|
|
Tmem47
|
transmembrane protein 47
|
|
chr9_+_45178485
|
14.961
|
|
Fxyd6
|
FXYD domain-containing ion transport regulator 6
|
|
chr9_-_60535699
|
14.958
|
|
Lrrc49
|
leucine rich repeat containing 49
|
|
chrX_+_5624820
|
14.920
|
|
Nudt11
|
nudix (nucleoside diphosphate linked moiety X)-type motif 11
|
|
chr5_+_34339351
|
14.913
|
|
Nat8l
|
N-acetyltransferase 8-like
|
|
chr18_-_74224541
|
14.895
|
NM_172632
|
Mapk4
|
mitogen-activated protein kinase 4
|
|
chr9_+_45178521
|
14.853
|
|
Fxyd6
|
FXYD domain-containing ion transport regulator 6
|
|
chr4_+_138011061
|
14.844
|
NM_025451
|
Camk2n1
|
calcium/calmodulin-dependent protein kinase II inhibitor 1
|
|
chr5_+_137729501
|
14.725
|
NM_009599
|
Ache
|
acetylcholinesterase
|
|
chr6_+_85978802
|
14.657
|
|
Add2
|
adducin 2 (beta)
|
|
chr5_+_34064372
|
14.626
|
NM_008010
NM_001163215
|
Fgfr3
|
fibroblast growth factor receptor 3
|
|
chr17_-_66799086
|
14.537
|
NM_001114098
NM_172963
|
1110012J17Rik
|
RIKEN cDNA 1110012J17 gene
|
|
chrX_-_71070284
|
14.512
|
NM_001029868
|
Pdzd4
|
PDZ domain containing 4
|
|
chr11_-_118770873
|
14.483
|
NM_001024931
NM_001039167
NM_001039168
|
Rbfox3
|
RNA binding protein, fox-1 homolog (C. elegans) 3
|
|
chr6_+_8899165
|
14.458
|
|
Nxph1
|
neurexophilin 1
|
|
chr11_-_5965743
|
14.439
|
NM_001174053
NM_001174054
NM_007595
|
Camk2b
|
calcium/calmodulin-dependent protein kinase II, beta
|
|
chr9_+_103014279
|
14.435
|
|
Rab6b
|
RAB6B, member RAS oncogene family
|
|
chr15_-_37721754
|
14.428
|
NM_001170866
|
Ncald
|
neurocalcin delta
|
|
chr11_-_118770859
|
14.403
|
|
Rbfox3
|
RNA binding protein, fox-1 homolog (C. elegans) 3
|
|
chr15_+_103333440
|
14.324
|
|
Pde1b
|
phosphodiesterase 1B, Ca2+-calmodulin dependent
|
|
chr6_-_127718746
|
14.321
|
NM_201371
|
Prmt8
|
protein arginine N-methyltransferase 8
|
|
chr7_+_64846558
|
14.092
|
|
Gabrb3
|
gamma-aminobutyric acid (GABA) A receptor, subunit beta 3
|
|
chr9_-_56008595
|
14.082
|
|
Tspan3
|
tetraspanin 3
|
|
chr4_+_129662284
|
14.049
|
NM_173071
|
Bai2
|
brain-specific angiogenesis inhibitor 2
|
|
chr9_-_56008819
|
13.956
|
|
Tspan3
|
tetraspanin 3
|
|
chr18_+_64425037
|
13.929
|
|
St8sia3
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3
|
|
chr8_+_98226914
|
13.883
|
NM_001195006
NM_145602
|
Ndrg4
|
N-myc downstream regulated gene 4
|
|
chr4_+_104040160
|
13.879
|
NM_010014
NM_177259
|
Dab1
|
disabled homolog 1 (Drosophila)
|
|
chr1_-_134638361
|
13.853
|
NM_001160317
NM_182716
|
Nfasc
|
neurofascin
|
|
chr11_-_5965544
|
13.844
|
|
Camk2b
|
calcium/calmodulin-dependent protein kinase II, beta
|
|
chr7_-_116925058
|
13.834
|
NM_020610
|
Nrip3
|
nuclear receptor interacting protein 3
|
|
chr4_+_137220675
|
13.829
|
|
Rap1gap
|
Rap1 GTPase-activating protein
|
|
chr9_+_57351848
|
13.813
|
NM_133982
|
Rpp25
|
ribonuclease P 25 subunit (human)
|
|
chr5_+_34338625
|
13.787
|
NM_001001985
|
Nat8l
|
N-acetyltransferase 8-like
|
|
chr9_-_56008770
|
13.766
|
|
Tspan3
|
tetraspanin 3
|
|
chr12_-_73337707
|
13.664
|
NM_001007596
|
Rtn1
|
reticulon 1
|
|
chr5_-_84846256
|
13.637
|
|
Epha5
|
Eph receptor A5
|
|
chr10_-_67375130
|
13.601
|
|
Zfp365
|
zinc finger protein 365
|
|
chr6_+_85978713
|
13.550
|
|
Add2
|
adducin 2 (beta)
|
|
chr9_+_103014463
|
13.461
|
|
Rab6b
|
RAB6B, member RAS oncogene family
|
|
chr11_-_64976408
|
13.409
|
NM_001099288
NM_175003
|
Arhgap44
|
Rho GTPase activating protein 44
|
|
chr13_+_54472806
|
13.372
|
|
Cplx2
|
complexin 2
|
|
chr15_-_99358387
|
13.348
|
NM_001038658
NM_028224
|
Faim2
|
Fas apoptotic inhibitory molecule 2
|
|
chr15_-_79635069
|
13.330
|
|
Nptxr
|
neuronal pentraxin receptor
|
|
chr15_+_99501148
|
13.249
|
NM_009597
|
Accn2
|
amiloride-sensitive cation channel 2, neuronal
|
|
chr2_+_132607432
|
13.229
|
|
Chgb
|
chromogranin B
|
|
chr6_-_13788453
|
13.198
|
|
Gpr85
|
G protein-coupled receptor 85
|
|
chr4_-_133053867
|
13.174
|
|
Trnp1
|
TMF1-regulated nuclear protein 1
|
|
chr15_-_98638348
|
13.022
|
NM_001013741
|
Ddn
|
dendrin
|
|
chr6_-_13788580
|
12.997
|
|
Gpr85
|
G protein-coupled receptor 85
|
|
chr4_-_124539414
|
12.983
|
NM_001007573
|
Maneal
|
mannosidase, endo-alpha-like
|
|
chr4_+_129662110
|
12.944
|
|
Bai2
|
brain-specific angiogenesis inhibitor 2
|
|
chr15_-_98638230
|
12.921
|
|
Ddn
|
dendrin
|
|
chr8_-_126383327
|
12.760
|
|
1700054N08Rik
|
RIKEN cDNA 1700054N08 gene
|
|
chr3_+_14886253
|
12.746
|
NM_009801
|
Car2
|
carbonic anhydrase 2
|
|
chr9_-_53823107
|
12.639
|
NM_177769
|
Elmod1
|
ELMO domain containing 1
|
|
chr12_-_71448300
|
12.620
|
|
Trim9
|
tripartite motif-containing 9
|
|
chr4_+_8462766
|
12.601
|
NM_021518
|
Rab2a
|
RAB2A, member RAS oncogene family
|
|
chr2_-_92232506
|
12.599
|
|
Mapk8ip1
|
mitogen-activated protein kinase 8 interacting protein 1
|
|
chr12_+_117724182
|
12.526
|
NM_011215
|
Ptprn2
|
protein tyrosine phosphatase, receptor type, N polypeptide 2
|
|
chr15_+_97938874
|
12.483
|
NM_001163487
|
Pfkm
|
phosphofructokinase, muscle
|
|
chr17_-_57199207
|
12.386
|
NM_025877
|
Slc25a23
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 23
|
|
chr14_+_14286471
|
12.234
|
NM_001163032
|
Synpr
|
synaptoporin
|
|
chr7_+_25266986
|
12.217
|
NM_153112
|
Cadm4
|
cell adhesion molecule 4
|
|
chr6_-_124813970
|
12.178
|
|
Gpr162
|
G protein-coupled receptor 162
|
|
chr2_+_132607408
|
12.149
|
|
Chgb
|
chromogranin B
|
|
chr6_+_29867012
|
12.135
|
NM_001037740
NM_177204
|
Fam40b
|
family with sequence similarity 40, member B
|
|
chr2_+_132607398
|
12.104
|
|
Chgb
|
chromogranin B
|
|
chr5_+_73882264
|
12.056
|
NM_001190733
NM_178896
|
Dcun1d4
|
DCN1, defective in cullin neddylation 1, domain containing 4 (S. cerevisiae)
|
|
chr10_+_13685882
|
12.019
|
|
Hivep2
|
human immunodeficiency virus type I enhancer binding protein 2
|
|
chr2_-_113669159
|
11.935
|
|
Scg5
|
secretogranin V
|
|
chr6_-_124813720
|
11.848
|
|
Gpr162
|
G protein-coupled receptor 162
|
|
chr2_-_152202301
|
11.826
|
NM_001009948
|
Nrsn2
|
neurensin 2
|
|
chr7_+_3303657
|
11.813
|
NM_011102
|
Prkcc
|
protein kinase C, gamma
|
|
chr10_-_33343965
|
11.668
|
|
Clvs2
|
clavesin 2
|
|
chr10_+_90038471
|
11.610
|
NM_001177397
|
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B
|
|
chr4_-_133054355
|
11.550
|
NM_001081156
|
Trnp1
|
TMF1-regulated nuclear protein 1
|
|
chr1_+_174271338
|
11.541
|
NM_001039484
|
Kcnj10
|
potassium inwardly-rectifying channel, subfamily J, member 10
|
|
chr11_+_29593657
|
11.512
|
NM_194051
|
Rtn4
|
reticulon 4
|
|
chr4_-_57002965
|
11.483
|
|
6430704M03Rik
|
RIKEN cDNA 6430704M03 gene
|
|
chr3_+_14886681
|
11.449
|
|
Car2
|
carbonic anhydrase 2
|
|
chr11_-_53293696
|
11.441
|
NM_183173
|
Ankrd43
|
ankyrin repeat domain 43
|
|
chr9_-_56008828
|
11.438
|
NM_019793
|
Tspan3
|
tetraspanin 3
|
|
chr11_-_53293580
|
11.427
|
|
Ankrd43
|
ankyrin repeat domain 43
|
|
chr3_-_107320881
|
11.406
|
NM_172271
|
Slc6a17
|
solute carrier family 6 (neurotransmitter transporter), member 17
|
|
chr2_-_113669243
|
11.401
|
NM_009162
|
Scg5
|
secretogranin V
|
|
chr8_-_67373608
|
11.389
|
|
Klhl2
|
kelch-like 2, Mayven (Drosophila)
|
|
chr13_-_36826245
|
11.384
|
NM_153529
|
Nrn1
|
neuritin 1
|
|
chr15_-_79635156
|
11.292
|
|
Nptxr
|
neuronal pentraxin receptor
|
|
chr8_+_121970660
|
11.286
|
|
Necab2
|
N-terminal EF-hand calcium binding protein 2
|
|
chr5_+_34339258
|
11.285
|
|
Nat8l
|
N-acetyltransferase 8-like
|
|
chr13_+_48356795
|
11.283
|
NM_031166
|
Id4
|
inhibitor of DNA binding 4
|
|
chr11_+_71564204
|
11.281
|
NM_177618
|
Wscd1
|
WSC domain containing 1
|
|
chr18_+_38115192
|
11.277
|
NM_153793
|
Rell2
|
RELT-like 2
|
|
chr11_-_5965555
|
11.275
|
|
Camk2b
|
calcium/calmodulin-dependent protein kinase II, beta
|
|
chr4_+_8462756
|
11.234
|
|
Rab2a
|
RAB2A, member RAS oncogene family
|
|
chr6_-_126595818
|
11.211
|
NM_010595
|
Kcna1
|
potassium voltage-gated channel, shaker-related subfamily, member 1
|
|
chr15_+_103333554
|
11.180
|
NM_008800
|
Pde1b
|
phosphodiesterase 1B, Ca2+-calmodulin dependent
|
|
chr2_-_180869947
|
11.098
|
|
Kcnq2
|
potassium voltage-gated channel, subfamily Q, member 2
|
|
chr2_-_118529608
|
11.040
|
NM_001081971
|
Gm1337
|
predicted gene 1337
|
|
chr9_+_103014374
|
11.023
|
NM_173781
|
Rab6b
|
RAB6B, member RAS oncogene family
|
|
chr2_-_127308234
|
11.009
|
NM_001111331
|
Kcnip3
|
Kv channel interacting protein 3, calsenilin
|
|
chr4_-_151235861
|
10.945
|
NM_001081557
NM_001195565
|
Camta1
|
calmodulin binding transcription activator 1
|
|
chr13_-_111070344
|
10.942
|
|
Rab3c
|
RAB3C, member RAS oncogene family
|
|
chr5_+_37633353
|
10.842
|
|
Crmp1
|
collapsin response mediator protein 1
|
|
chr4_-_149148173
|
10.838
|
NM_027460
|
Slc25a33
|
solute carrier family 25, member 33
|
|
chr14_+_71289902
|
10.825
|
|
Gfra2
|
glial cell line derived neurotrophic factor family receptor alpha 2
|
|
chr7_+_3303770
|
10.819
|
|
Prkcc
|
protein kinase C, gamma
|
|
chr17_+_86567003
|
10.754
|
NM_011104
|
Prkce
|
protein kinase C, epsilon
|
|
chr19_+_6427922
|
10.724
|
NM_020253
|
Nrxn2
|
neurexin II
|
|
chr4_+_126846429
|
10.696
|
NM_198618
|
Dlgap3
|
discs, large (Drosophila) homolog-associated protein 3
|
|
chr15_-_37722161
|
10.599
|
NM_134094
|
Ncald
|
neurocalcin delta
|
|
chr6_+_49772688
|
10.528
|
NM_023456
|
Npy
|
neuropeptide Y
|
|
chr9_-_69989285
|
10.500
|
NM_029784
|
Fam81a
|
family with sequence similarity 81, member A
|
|
chr10_-_80945445
|
10.438
|
NM_176954
|
Celf5
|
CUGBP, Elav-like family member 5
|
|
chr16_-_32246968
|
10.434
|
NM_173439
|
Fbxo45
|
F-box protein 45
|
|
chr11_+_55017825
|
10.426
|
NM_153139
|
Slc36a1
|
solute carrier family 36 (proton/amino acid symporter), member 1
|
|
chr12_-_73337534
|
10.422
|
|
Rtn1
|
reticulon 1
|
|
chr8_+_107872157
|
10.413
|
NM_001081332
|
Slc9a5
|
solute carrier family 9 (sodium/hydrogen exchanger), member 5
|
|
chr5_+_88994386
|
10.372
|
|
Rufy3
|
RUN and FYVE domain containing 3
|
|
chr1_+_75479559
|
10.372
|
|
Tmem198
|
transmembrane protein 198
|
|
chr7_+_125856066
|
10.346
|
NM_182995
|
6330503K22Rik
|
RIKEN cDNA 6330503K22 gene
|
|
chr2_-_91284827
|
10.344
|
|
1110051M20Rik
|
RIKEN cDNA 1110051M20 gene
|
|
chr11_-_115128586
|
10.257
|
|
Grin2c
|
glutamate receptor, ionotropic, NMDA2C (epsilon 3)
|
|
chr10_+_78243274
|
10.256
|
|
Slc1a6
|
solute carrier family 1 (high affinity aspartate/glutamate transporter), member 6
|
|
chr2_+_135883662
|
10.254
|
NM_029530
|
6330527O06Rik
|
RIKEN cDNA 6330527O06 gene
|
|
chr3_-_117063317
|
10.226
|
|
D3Bwg0562e
|
DNA segment, Chr 3, Brigham & Women's Genetics 0562 expressed
|
|
chr11_-_35612058
|
10.183
|
|
Fbll1
|
fibrillarin-like 1
|
|
chr14_+_56129284
|
10.162
|
NM_001136057
NM_001146183
|
Cpne6
|
copine VI
|