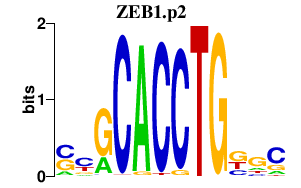
|
chr7_-_30066910
|
97.441
|
NM_001082548
NM_011464
|
Spint2
|
serine protease inhibitor, Kunitz type 2
|
|
chr7_-_30066867
|
95.781
|
|
Spint2
|
serine protease inhibitor, Kunitz type 2
|
|
chr7_-_30066642
|
90.330
|
|
Spint2
|
serine protease inhibitor, Kunitz type 2
|
|
chr15_+_54402988
|
59.521
|
|
Mal2
|
mal, T-cell differentiation protein 2
|
|
chr12_-_74647339
|
57.902
|
NM_178715
|
Tmem30b
|
transmembrane protein 30B
|
|
chr8_+_109127245
|
52.823
|
NM_009864
|
Cdh1
|
cadherin 1
|
|
chr11_+_69778998
|
51.535
|
|
Cldn7
|
claudin 7
|
|
chr15_+_54402914
|
51.138
|
NM_178920
|
Mal2
|
mal, T-cell differentiation protein 2
|
|
chr8_+_3587791
|
45.179
|
|
2310057J16Rik
|
RIKEN cDNA 2310057J16 gene
|
|
chr11_+_69778907
|
43.492
|
NM_016887
|
Cldn7
|
claudin 7
|
|
chr4_-_11313886
|
40.391
|
NM_194055
|
Esrp1
|
epithelial splicing regulatory protein 1
|
|
chr8_+_3587675
|
37.133
|
|
2310057J16Rik
|
RIKEN cDNA 2310057J16 gene
|
|
chr16_+_91270319
|
36.383
|
|
Olig1
|
oligodendrocyte transcription factor 1
|
|
chr17_+_88036128
|
34.834
|
|
Epcam
|
epithelial cell adhesion molecule
|
|
chr11_-_35793590
|
34.262
|
NM_170779
|
Wwc1
|
WW, C2 and coiled-coil domain containing 1
|
|
chr7_-_31758390
|
33.748
|
NM_001164184
NM_001164185
NM_017405
|
Lsr
|
lipolysis stimulated lipoprotein receptor
|
|
chrX_+_162676903
|
33.249
|
|
Gpm6b
|
glycoprotein m6b
|
|
chr17_+_57201623
|
32.778
|
NM_177638
|
Crb3
|
crumbs homolog 3 (Drosophila)
|
|
chr1_+_194979305
|
30.491
|
NM_016851
|
Irf6
|
interferon regulatory factor 6
|
|
chr8_-_41720091
|
30.289
|
NM_001040699
|
Mtmr7
|
myotubularin related protein 7
|
|
chrX_+_162676870
|
30.104
|
NM_001177961
NM_001177962
NM_023122
|
Gpm6b
|
glycoprotein m6b
|
|
chr2_+_25258568
|
30.037
|
NM_008721
|
Npdc1
|
neural proliferation, differentiation and control gene 1
|
|
chr2_+_25258653
|
29.061
|
|
Npdc1
|
neural proliferation, differentiation and control gene 1
|
|
chr3_-_55987612
|
28.486
|
NM_030595
|
Nbea
|
neurobeachin
|
|
chr18_+_74602260
|
27.963
|
NM_201600
|
Myo5b
|
myosin VB
|
|
chr11_-_105805442
|
27.121
|
NM_007805
|
Cyb561
|
cytochrome b-561
|
|
chr8_+_3587449
|
26.616
|
NM_001163749
NM_027171
|
2310057J16Rik
|
RIKEN cDNA 2310057J16 gene
|
|
chr8_+_12915892
|
26.082
|
NM_178076
|
Mcf2l
|
mcf.2 transforming sequence-like
|
|
chr7_-_31758185
|
25.608
|
|
Lsr
|
lipolysis stimulated lipoprotein receptor
|
|
chr2_+_90730089
|
25.532
|
|
C1qtnf4
|
C1q and tumor necrosis factor related protein 4
|
|
chr1_+_120524521
|
25.488
|
NM_023755
|
Tcfcp2l1
|
transcription factor CP2-like 1
|
|
chr17_+_88035996
|
25.442
|
|
Epcam
|
epithelial cell adhesion molecule
|
|
chr13_-_101322557
|
25.207
|
|
Ocln
|
occludin
|
|
chr1_-_79855209
|
25.007
|
|
Serpine2
|
serine (or cysteine) peptidase inhibitor, clade E, member 2
|
|
chr13_-_101322672
|
24.561
|
|
Ocln
|
occludin
|
|
chr10_-_18463500
|
24.371
|
NM_001033258
|
D10Bwg1379e
|
DNA segment, Chr 10, Brigham & Women's Genetics 1379 expressed
|
|
chr16_+_91270009
|
22.736
|
NM_016968
|
Olig1
|
oligodendrocyte transcription factor 1
|
|
chr4_+_133074877
|
22.479
|
NM_001083916
NM_133707
|
1810019J16Rik
|
RIKEN cDNA 1810019J16 gene
|
|
chr12_+_113960424
|
22.282
|
|
AW555464
|
expressed sequence AW555464
|
|
chr18_+_20716598
|
22.197
|
NM_007883
|
Dsg2
|
desmoglein 2
|
|
chr8_+_86424565
|
22.178
|
|
Lphn1
|
latrophilin 1
|
|
chr12_+_113960530
|
22.143
|
|
AW555464
|
expressed sequence AW555464
|
|
chr13_-_101322416
|
21.555
|
NM_008756
|
Ocln
|
occludin
|
|
chr4_+_137263376
|
21.517
|
|
Rap1gap
|
Rap1 GTPase-activating protein
|
|
chr1_+_78306890
|
21.350
|
NM_001004173
|
Sgpp2
|
sphingosine-1-phosphate phosphotase 2
|
|
chr2_+_119063090
|
21.337
|
NM_016907
|
Spint1
|
serine protease inhibitor, Kunitz type 1
|
|
chr7_-_20281956
|
21.153
|
|
|
|
|
chr2_+_119063134
|
20.363
|
|
Spint1
|
serine protease inhibitor, Kunitz type 1
|
|
chr4_+_133074976
|
20.259
|
|
1810019J16Rik
|
RIKEN cDNA 1810019J16 gene
|
|
chr11_-_100259048
|
20.250
|
NM_010593
|
Jup
|
junction plakoglobin
|
|
chr2_+_119063194
|
20.217
|
|
|
|
|
chr15_-_78633301
|
20.184
|
NM_130859
|
Card10
|
caspase recruitment domain family, member 10
|
|
chr14_+_30822097
|
20.084
|
NM_001136240
|
Chdh
|
choline dehydrogenase
|
|
chr11_+_115685417
|
20.002
|
|
Llgl2
|
lethal giant larvae homolog 2 (Drosophila)
|
|
chr1_-_79855229
|
19.971
|
NM_009255
|
Serpine2
|
serine (or cysteine) peptidase inhibitor, clade E, member 2
|
|
chr1_-_79855177
|
19.918
|
|
Serpine2
|
serine (or cysteine) peptidase inhibitor, clade E, member 2
|
|
chr19_+_6306456
|
19.885
|
NM_001033342
|
Cdc42bpg
|
CDC42 binding protein kinase gamma (DMPK-like)
|
|
chr17_+_88036047
|
19.865
|
|
Epcam
|
epithelial cell adhesion molecule
|
|
chr16_-_97985323
|
19.158
|
NM_023663
|
Ripk4
|
receptor-interacting serine-threonine kinase 4
|
|
chr6_+_4853319
|
18.821
|
NM_181595
|
Ppp1r9a
|
protein phosphatase 1, regulatory (inhibitor) subunit 9A
|
|
chr9_-_95745663
|
18.790
|
NM_001033210
|
Pls1
|
plastin 1 (I-isoform)
|
|
chr11_+_115685365
|
18.695
|
NM_145438
|
Llgl2
|
lethal giant larvae homolog 2 (Drosophila)
|
|
chr4_-_151235861
|
18.494
|
NM_001081557
NM_001195565
|
Camta1
|
calmodulin binding transcription activator 1
|
|
chr8_-_108660711
|
18.288
|
|
Esrp2
|
epithelial splicing regulatory protein 2
|
|
chr13_+_97841524
|
18.021
|
NM_001100458
|
Fam169a
|
family with sequence similarity 169, member A
|
|
chr16_+_16213397
|
17.825
|
|
Pkp2
|
plakophilin 2
|
|
chr5_+_37637027
|
17.727
|
NM_007765
|
Crmp1
|
collapsin response mediator protein 1
|
|
chrX_+_97972559
|
17.531
|
NM_001177780
|
Dlg3
|
discs, large homolog 3 (Drosophila)
|
|
chr5_-_44492908
|
17.508
|
NM_001163577
NM_008935
|
Prom1
|
prominin 1
|
|
chr3_+_121129396
|
17.418
|
NM_028044
|
Cnn3
|
calponin 3, acidic
|
|
chr14_-_71035662
|
17.355
|
|
Epb4.9
|
erythrocyte protein band 4.9
|
|
chr8_+_109127326
|
17.282
|
|
Cdh1
|
cadherin 1
|
|
chr5_-_17866110
|
17.247
|
NM_010305
|
Gnai1
|
guanine nucleotide binding protein (G protein), alpha inhibiting 1
|
|
chr2_+_90729768
|
17.086
|
|
C1qtnf4
|
C1q and tumor necrosis factor related protein 4
|
|
chr4_-_133794246
|
16.735
|
NM_001081047
|
Cnksr1
|
connector enhancer of kinase suppressor of Ras 1
|
|
chr14_+_102129129
|
16.598
|
NM_201529
|
Lmo7
|
LIM domain only 7
|
|
chr16_+_16213434
|
16.580
|
NM_026163
|
Pkp2
|
plakophilin 2
|
|
chr8_-_108660736
|
16.286
|
NM_176838
|
Esrp2
|
epithelial splicing regulatory protein 2
|
|
chr2_+_126378563
|
15.917
|
NM_011978
|
Slc27a2
|
solute carrier family 27 (fatty acid transporter), member 2
|
|
chr1_-_89406861
|
15.696
|
NM_019867
|
Ngef
|
neuronal guanine nucleotide exchange factor
|
|
chr17_+_28906259
|
15.634
|
NM_011950
|
Mapk13
|
mitogen-activated protein kinase 13
|
|
chr11_-_20731016
|
15.566
|
NM_173752
|
1110067D22Rik
|
RIKEN cDNA 1110067D22 gene
|
|
chr2_+_167906462
|
15.318
|
NM_021409
|
Pard6b
|
par-6 (partitioning defective 6) homolog beta (C. elegans)
|
|
chr19_-_36811137
|
15.155
|
|
Ppp1r3c
|
protein phosphatase 1, regulatory (inhibitor) subunit 3C
|
|
chr7_+_148401781
|
15.130
|
|
Rassf7
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7
|
|
chr2_-_37558794
|
15.035
|
|
Strbp
|
spermatid perinuclear RNA binding protein
|
|
chr9_-_29219306
|
14.986
|
|
Ntm
|
neurotrimin
|
|
chr14_-_8491067
|
14.972
|
|
Gm2897
|
predicted gene 2897
|
|
chr19_-_36811090
|
14.868
|
NM_016854
|
Ppp1r3c
|
protein phosphatase 1, regulatory (inhibitor) subunit 3C
|
|
chr9_+_36640268
|
14.863
|
|
Fez1
|
fasciculation and elongation protein zeta 1 (zygin I)
|
|
chr4_+_133074949
|
14.748
|
|
1810019J16Rik
|
RIKEN cDNA 1810019J16 gene
|
|
chr8_-_112486020
|
14.730
|
NM_028584
NM_212447
|
Marveld3
|
MARVEL (membrane-associating) domain containing 3
|
|
chr11_-_115229039
|
14.711
|
|
C630004H02Rik
|
RIKEN cDNA C630004H02 gene
|
|
chr8_-_113917651
|
14.270
|
NM_178086
|
Fa2h
|
fatty acid 2-hydroxylase
|
|
chr1_+_137715174
|
14.266
|
NM_133664
|
Lad1
|
ladinin
|
|
chr13_+_38243146
|
14.145
|
NM_023842
|
Dsp
|
desmoplakin
|
|
chr9_-_29219382
|
14.024
|
|
Ntm
|
neurotrimin
|
|
chr11_-_100259061
|
13.983
|
|
Jup
|
junction plakoglobin
|
|
chr4_+_95634005
|
13.960
|
NM_030014
|
Hook1
|
hook homolog 1 (Drosophila)
|
|
chr6_+_88674398
|
13.863
|
NM_001166249
NM_011844
|
Mgll
|
monoglyceride lipase
|
|
chr13_-_34437042
|
13.706
|
NM_001033167
|
Slc22a23
|
solute carrier family 22, member 23
|
|
chr15_-_75839859
|
13.339
|
NM_001168253
|
Fam83h
|
family with sequence similarity 83, member H
|
|
chrX_+_5624624
|
13.327
|
NM_021431
|
Nudt11
|
nudix (nucleoside diphosphate linked moiety X)-type motif 11
|
|
chr7_-_106331080
|
13.306
|
NM_026384
|
Dgat2
|
diacylglycerol O-acyltransferase 2
|
|
chr6_-_134582365
|
13.255
|
NM_026345
|
Mansc1
|
MANSC domain containing 1
|
|
chrX_+_5624820
|
13.118
|
|
Nudt11
|
nudix (nucleoside diphosphate linked moiety X)-type motif 11
|
|
chr4_+_84851330
|
12.924
|
NM_019535
|
Sh3gl2
|
SH3-domain GRB2-like 2
|
|
chr3_-_57651621
|
12.898
|
NM_019410
|
Pfn2
|
profilin 2
|
|
chr11_-_35793951
|
12.869
|
|
Wwc1
|
WW, C2 and coiled-coil domain containing 1
|
|
chr1_-_166388431
|
12.799
|
NM_009721
|
Atp1b1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide
|
|
chr9_+_120213190
|
12.764
|
NM_144557
|
Myrip
|
myosin VIIA and Rab interacting protein
|
|
chr7_-_4741145
|
12.664
|
NM_029384
|
2210411K11Rik
|
RIKEN cDNA 2210411K11 gene
|
|
chr12_+_114378368
|
12.514
|
NM_024223
|
Crip2
|
cysteine rich protein 2
|
|
chr16_-_18629938
|
12.404
|
NM_213614
|
Sept5
|
septin 5
|
|
chr5_+_135462067
|
12.388
|
NM_009902
|
Cldn3
|
claudin 3
|
|
chrX_+_155852439
|
12.326
|
NM_001081124
|
Mtap7d2
|
MAP7 domain containing 2
|
|
chr14_-_7937970
|
12.299
|
NM_001024712
|
Gm3696
|
predicted gene 3696
|
|
chr6_+_48487648
|
12.016
|
|
Atp6v0e2
|
ATPase, H+ transporting, lysosomal V0 subunit E2
|
|
chr4_-_129952771
|
12.007
|
NM_172702
|
Serinc2
|
serine incorporator 2
|
|
chr19_-_24299471
|
11.998
|
NM_011597
|
Tjp2
|
tight junction protein 2
|
|
chr14_+_3049284
|
11.949
|
NM_001177715
NM_001177714
|
Gm2897
|
predicted gene 2897
|
|
chr13_-_62989500
|
11.930
|
|
Fbp1
|
fructose bisphosphatase 1
|
|
chr4_+_104040160
|
11.924
|
NM_010014
NM_177259
|
Dab1
|
disabled homolog 1 (Drosophila)
|
|
chr4_+_43071801
|
11.754
|
|
Unc13b
|
unc-13 homolog B (C. elegans)
|
|
chr5_-_140290195
|
11.721
|
NM_001161548
|
Tmem184a
|
transmembrane protein 184a
|
|
chr2_-_148558110
|
11.662
|
NM_019632
|
Napb
|
N-ethylmaleimide sensitive fusion protein attachment protein beta
|
|
chr6_+_48487567
|
11.588
|
NM_133764
|
Atp6v0e2
|
ATPase, H+ transporting, lysosomal V0 subunit E2
|
|
chr15_-_34608458
|
11.576
|
NM_145469
|
Nipal2
|
NIPA-like domain containing 2
|
|
chr4_+_101169252
|
11.575
|
NM_198412
|
Dnajc6
|
DnaJ (Hsp40) homolog, subfamily C, member 6
|
|
chr8_-_4259126
|
11.541
|
NM_183315
|
Ctxn1
|
cortexin 1
|
|
chr11_-_83881811
|
11.495
|
NM_019819
|
Dusp14
|
dual specificity phosphatase 14
|
|
chr14_-_7859962
|
11.465
|
NM_001164727
|
Gm10406
|
predicted gene 10406
|
|
chr11_-_80593259
|
11.202
|
NM_177390
|
Myo1d
|
myosin ID
|
|
chr6_+_91106773
|
11.125
|
NM_144919
|
Hdac11
|
histone deacetylase 11
|
|
chr12_+_80130149
|
11.071
|
NM_181073
|
Plekhh1
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 1
|
|
chr5_+_124423632
|
10.985
|
NM_145070
|
Hip1r
|
huntingtin interacting protein 1 related
|
|
chr9_-_21397188
|
10.846
|
NM_138303
|
Yipf2
|
Yip1 domain family, member 2
|
|
chr19_-_24299389
|
10.820
|
|
Tjp2
|
tight junction protein 2
|
|
chr8_-_63461923
|
10.810
|
|
Clcn3
|
chloride channel 3
|
|
chr12_+_85350438
|
10.803
|
NM_012006
|
Acot1
|
acyl-CoA thioesterase 1
|
|
chr7_+_148401731
|
10.738
|
NM_025886
|
Rassf7
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7
|
|
chr4_+_148960536
|
10.710
|
|
Clstn1
|
calsyntenin 1
|
|
chr2_+_138082301
|
10.543
|
NM_001025431
|
Btbd3
|
BTB (POZ) domain containing 3
|
|
chr12_-_80007923
|
10.542
|
NM_013738
|
Plek2
|
pleckstrin 2
|
|
chr12_+_113960379
|
10.452
|
NM_001024602
|
AW555464
|
expressed sequence AW555464
|
|
chr6_-_92656148
|
10.393
|
NM_001081146
|
Prickle2
|
prickle homolog 2 (Drosophila)
|
|
chr3_+_62142698
|
10.379
|
NM_001081295
|
Arhgef26
|
Rho guanine nucleotide exchange factor (GEF) 26
|
|
chr10_+_61111426
|
10.354
|
|
Ppa1
|
pyrophosphatase (inorganic) 1
|
|
chr17_+_87153083
|
10.351
|
NM_010137
|
Epas1
|
endothelial PAS domain protein 1
|
|
chr11_+_98308333
|
10.333
|
|
Grb7
|
growth factor receptor bound protein 7
|
|
chrX_-_132749976
|
10.291
|
NM_009052
|
Bex1
|
brain expressed gene 1
|
|
chr6_-_137118238
|
10.277
|
NM_001164212
NM_001164214
|
Rerg
|
RAS-like, estrogen-regulated, growth-inhibitor
|
|
chr12_-_58646914
|
10.248
|
|
Foxa1
|
forkhead box A1
|
|
chr12_-_58647074
|
10.137
|
NM_008259
|
Foxa1
|
forkhead box A1
|
|
chr4_-_153673990
|
10.135
|
NM_001112744
|
Arhgef16
|
Rho guanine nucleotide exchange factor (GEF) 16
|
|
chr7_+_108470264
|
10.120
|
|
Stard10
|
START domain containing 10
|
|
chr1_+_137715339
|
10.074
|
|
Lad1
|
ladinin
|
|
chr3_-_88351996
|
10.024
|
NM_016899
|
Rab25
|
RAB25, member RAS oncogene family
|
|
chr5_-_145118920
|
10.018
|
NM_025833
|
Baiap2l1
|
BAI1-associated protein 2-like 1
|
|
chr7_-_3629111
|
9.998
|
NM_181820
|
Tmc4
|
transmembrane channel-like gene family 4
|
|
chr6_-_137118197
|
9.983
|
|
Rerg
|
RAS-like, estrogen-regulated, growth-inhibitor
|
|
chr9_+_69137659
|
9.905
|
|
Rora
|
RAR-related orphan receptor alpha
|
|
chr13_-_25029458
|
9.881
|
NM_172532
|
Aldh5a1
|
aldhehyde dehydrogenase family 5, subfamily A1
|
|
chr3_-_94590356
|
9.813
|
NM_001037711
|
Cgn
|
cingulin
|
|
chr18_-_34166859
|
9.787
|
NM_013512
|
Epb4.1l4a
|
erythrocyte protein band 4.1-like 4a
|
|
chrX_-_70905384
|
9.763
|
|
Pnck
|
pregnancy upregulated non-ubiquitously expressed CaM kinase
|
|
chr7_-_3629066
|
9.718
|
|
Tmc4
|
transmembrane channel-like gene family 4
|
|
chr5_-_63157309
|
9.636
|
NM_178407
|
Arap2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2
|
|
chr8_-_92872139
|
9.628
|
NM_172913
|
Tox3
|
TOX high mobility group box family member 3
|
|
chr14_-_122196929
|
9.621
|
NM_134074
|
Dock9
|
dedicator of cytokinesis 9
|
|
chr14_-_57723453
|
9.603
|
|
Gjb2
|
gap junction protein, beta 2
|
|
chr7_-_51920870
|
9.600
|
NM_028021
|
Myh14
|
myosin, heavy polypeptide 14
|
|
chr1_-_155033576
|
9.598
|
NM_008485
|
Lamc2
|
laminin, gamma 2
|
|
chr17_+_43938789
|
9.588
|
NM_207649
|
Rcan2
|
regulator of calcineurin 2
|
|
chr16_-_18089085
|
9.535
|
NM_011172
|
Prodh
|
proline dehydrogenase
|
|
chr10_+_61111331
|
9.529
|
NM_026438
|
Ppa1
|
pyrophosphatase (inorganic) 1
|
|
chr13_-_62989520
|
9.519
|
|
Fbp1
|
fructose bisphosphatase 1
|
|
chr5_+_124423712
|
9.516
|
|
Hip1r
|
huntingtin interacting protein 1 related
|
|
chr15_+_38837818
|
9.473
|
NM_001162494
|
Fzd6
|
frizzled homolog 6 (Drosophila)
|
|
chr15_+_38837839
|
9.450
|
NM_008056
|
Fzd6
|
frizzled homolog 6 (Drosophila)
|
|
chr4_+_43071852
|
9.449
|
NM_001081413
NM_021468
|
Unc13b
|
unc-13 homolog B (C. elegans)
|
|
chr15_-_37721754
|
9.423
|
NM_001170866
|
Ncald
|
neurocalcin delta
|
|
chr2_+_49474840
|
9.392
|
|
Kif5c
|
kinesin family member 5C
|
|
chr10_-_79600131
|
9.373
|
NM_001195268
|
Dos
|
downstream of Stk11
|
|
chr7_+_36119340
|
9.358
|
|
Rhpn2
|
rhophilin, Rho GTPase binding protein 2
|
|
chr4_+_129662110
|
9.347
|
|
Bai2
|
brain-specific angiogenesis inhibitor 2
|
|
chr11_-_12364886
|
9.337
|
NM_172496
|
Cobl
|
cordon-bleu
|
|
chr7_+_36119255
|
9.334
|
NM_027897
|
Rhpn2
|
rhophilin, Rho GTPase binding protein 2
|
|
chr7_+_17529961
|
9.324
|
NM_001099636
|
Pnmal2
|
PNMA-like 2
|
|
chr5_+_148768875
|
9.213
|
NM_029920
|
Mtus2
|
microtubule associated tumor suppressor candidate 2
|
|
chr11_-_94938114
|
9.174
|
|
Itga3
|
integrin alpha 3
|
|
chr9_-_99258896
|
9.130
|
NM_177775
|
Esyt3
|
extended synaptotagmin-like protein 3
|
|
chr15_-_37722161
|
9.123
|
NM_134094
|
Ncald
|
neurocalcin delta
|
|
chr11_-_100007218
|
9.121
|
NM_008471
|
Krt19
|
keratin 19
|
|
chr17_-_35838974
|
9.086
|
NM_001198831
|
Ddr1
|
discoidin domain receptor family, member 1
|
|
chr8_-_48374715
|
9.066
|
|
Stox2
|
storkhead box 2
|
|
chr9_+_108997946
|
9.065
|
NM_172775
|
Plxnb1
|
plexin B1
|
|
chr16_-_85173764
|
9.034
|
|
App
|
amyloid beta (A4) precursor protein
|
|
chr3_-_81878554
|
9.018
|
NM_001161796
NM_017469
|
Gucy1b3
|
guanylate cyclase 1, soluble, beta 3
|
|
chr9_+_69137694
|
8.939
|
|
Rora
|
RAR-related orphan receptor alpha
|