|
chr11_+_104092809
|
65.217
|
|
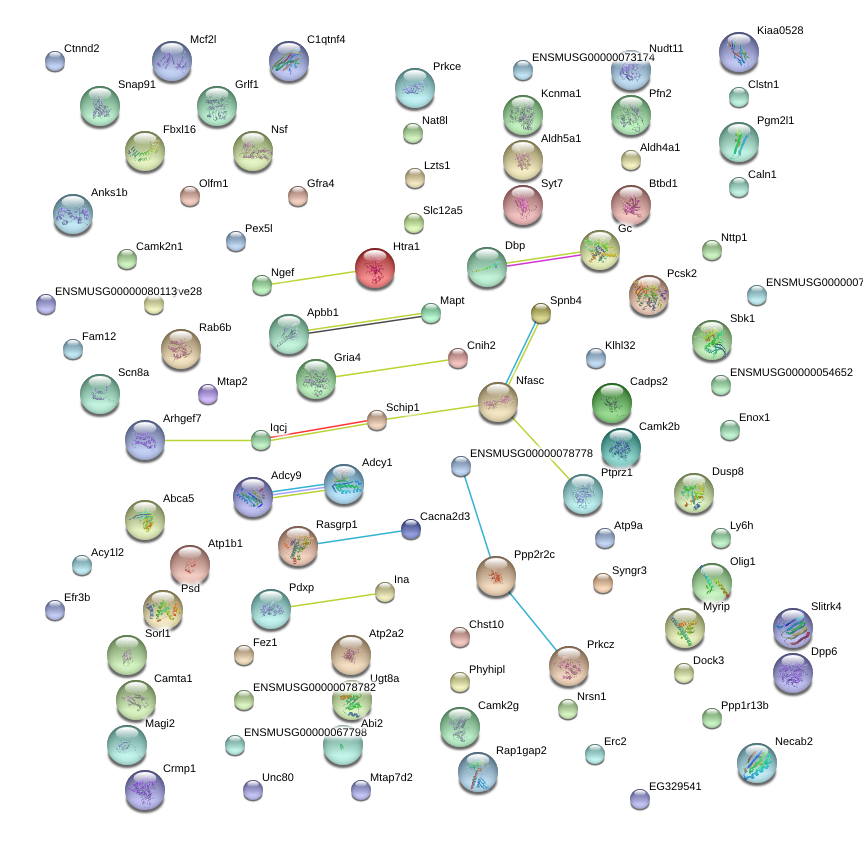
Mapt
|
microtubule-associated protein tau
|
|
chr11_+_104092793
|
61.994
|
|
Mapt
|
microtubule-associated protein tau
|
|
chr19_+_47089736
|
59.796
|
|
Ina
|
internexin neuronal intermediate filament protein, alpha
|
|
chr2_-_168567422
|
58.361
|
|
Atp9a
|
ATPase, class II, type 9A
|
|
chr7_-_149280946
|
52.942
|
|
Dusp8
|
dual specificity phosphatase 8
|
|
chr11_-_103815291
|
52.730
|
|
Nsf
|
N-ethylmaleimide sensitive fusion protein
|
|
chrX_-_61530035
|
50.310
|
|
Slitrk4
|
SLIT and NTRK-like family, member 4
|
|
chr7_-_112730004
|
50.029
|
NM_009685
|
Apbb1
|
amyloid beta (A4) precursor protein-binding, family B, member 1
|
|
chr2_-_168567436
|
49.712
|
|
Atp9a
|
ATPase, class II, type 9A
|
|
chr14_+_28435613
|
49.171
|
NM_177814
|
Erc2
|
ELKS/RAB6-interacting/CAST family member 2
|
|
chr11_-_103815361
|
46.411
|
NM_008740
|
Nsf
|
N-ethylmaleimide sensitive fusion protein
|
|
chr2_-_168567602
|
44.896
|
|
Atp9a
|
ATPase, class II, type 9A
|
|
chr7_-_28181562
|
43.906
|
|
Spnb4
|
spectrin beta 4
|
|
chr17_+_25945986
|
43.772
|
NM_001164225
|
Fbxl16
|
F-box and leucine-rich repeat protein 16
|
|
chr1_-_89406861
|
43.316
|
NM_019867
|
Ngef
|
neuronal guanine nucleotide exchange factor
|
|
chr9_+_103014279
|
43.266
|
|
Rab6b
|
RAB6B, member RAS oncogene family
|
|
chr9_-_86775154
|
43.028
|
NM_013669
|
Snap91
|
synaptosomal-associated protein 91
|
|
chr11_-_103815317
|
42.819
|
|
Nsf
|
N-ethylmaleimide sensitive fusion protein
|
|
chr4_-_151235861
|
42.734
|
NM_001081557
NM_001195565
|
Camta1
|
calmodulin binding transcription activator 1
|
|
chr16_+_5885447
|
42.391
|
|
Rbfox1
|
RNA binding protein, fox-1 homolog (C. elegans) 1
|
|
chrX_+_155852439
|
41.803
|
NM_001081124
|
Mtap7d2
|
MAP7 domain containing 2
|
|
chr9_+_36640268
|
41.092
|
|
Fez1
|
fasciculation and elongation protein zeta 1 (zygin I)
|
|
chr2_+_143371807
|
40.979
|
NM_008792
|
Pcsk2
|
proprotein convertase subtilisin/kexin type 2
|
|
chrX_-_61530170
|
40.390
|
NM_178740
|
Slitrk4
|
SLIT and NTRK-like family, member 4
|
|
chrX_+_155852641
|
40.376
|
|
Mtap7d2
|
MAP7 domain containing 2
|
|
chr4_-_150503464
|
39.559
|
|
Camta1
|
calmodulin binding transcription activator 1
|
|
chr11_+_104092749
|
38.671
|
NM_001038609
NM_010838
|
Mapt
|
microtubule-associated protein tau
|
|
chr9_-_107134182
|
38.141
|
NM_153413
|
Dock3
|
dedicator of cyto-kinesis 3
|
|
chr11_-_74403603
|
38.104
|
NM_001015046
|
Rap1gap2
|
RAP1 GTPase activating protein 2
|
|
chr7_-_112729716
|
37.195
|
|
Apbb1
|
amyloid beta (A4) precursor protein-binding, family B, member 1
|
|
chr5_+_34339351
|
37.180
|
|
Nat8l
|
N-acetyltransferase 8-like
|
|
chr3_-_57651621
|
36.740
|
NM_019410
|
Pfn2
|
profilin 2
|
|
chr5_+_34339258
|
36.492
|
|
Nat8l
|
N-acetyltransferase 8-like
|
|
chr11_-_5965743
|
36.025
|
NM_001174053
NM_001174054
NM_007595
|
Camk2b
|
calcium/calmodulin-dependent protein kinase II, beta
|
|
chr2_+_90730089
|
35.544
|
|
C1qtnf4
|
C1q and tumor necrosis factor related protein 4
|
|
chr5_+_130924661
|
34.990
|
NM_021371
|
Caln1
|
calneuron 1
|
|
chr16_+_6349108
|
34.941
|
|
Rbfox1
|
RNA binding protein, fox-1 homolog (C. elegans) 1
|
|
chr5_+_34339551
|
34.898
|
|
Nat8l
|
N-acetyltransferase 8-like
|
|
chr1_-_134638361
|
34.340
|
NM_001160317
NM_182716
|
Nfasc
|
neurofascin
|
|
chr2_-_117168600
|
33.776
|
NM_011246
|
Rasgrp1
|
RAS guanyl releasing protein 1
|
|
chr11_+_6963421
|
33.587
|
NM_009622
|
Adcy1
|
adenylate cyclase 1
|
|
chr3_-_33042050
|
32.650
|
|
Pex5l
|
peroxisomal biogenesis factor 5-like
|
|
chr7_+_107376116
|
32.574
|
NM_027629
|
Pgm2l1
|
phosphoglucomutase 2-like 1
|
|
chr15_+_30102306
|
32.301
|
NM_008729
|
Ctnnd2
|
catenin (cadherin associated protein), delta 2
|
|
chr8_+_12915892
|
32.248
|
NM_178076
|
Mcf2l
|
mcf.2 transforming sequence-like
|
|
chr8_+_121970660
|
32.043
|
|
Necab2
|
N-terminal EF-hand calcium binding protein 2
|
|
chr19_-_5098486
|
31.714
|
|
Cnih2
|
cornichon homolog 2 (Drosophila)
|
|
chr17_-_24826812
|
31.692
|
|
Syngr3
|
synaptogyrin 3
|
|
chr9_-_41932371
|
31.569
|
NM_011436
|
Sorl1
|
sortilin-related receptor, LDLR class A repeats-containing
|
|
chr4_-_154735351
|
31.410
|
NM_008860
|
Prkcz
|
protein kinase C, zeta
|
|
chr19_+_10463540
|
30.841
|
NM_018801
NM_173067
NM_173068
|
Syt7
|
synaptotagmin VII
|
|
chr9_+_120213190
|
30.747
|
NM_144557
|
Myrip
|
myosin VIIA and Rab interacting protein
|
|
chr1_-_166387998
|
30.604
|
|
Atp1b1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide
|
|
chr14_+_77556622
|
30.412
|
NM_172813
|
Enox1
|
ecto-NOX disulfide-thiol exchanger 1
|
|
chr10_+_90292388
|
30.377
|
|
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B
|
|
chr1_-_37776633
|
30.256
|
NM_028096
|
2010300C02Rik
|
RIKEN cDNA 2010300C02 gene
|
|
chr4_-_24778203
|
30.068
|
NM_001033531
NM_001163020
|
Klhl32
|
kelch-like 32 (Drosophila)
|
|
chr19_-_46401618
|
30.036
|
NM_028627
|
Psd
|
pleckstrin and Sec7 domain containing
|
|
chr1_+_66515003
|
30.025
|
NM_175510
|
Unc80
|
unc-80 homolog (C. elegans)
|
|
chr9_+_103014463
|
29.721
|
|
Rab6b
|
RAB6B, member RAS oncogene family
|
|
chr9_-_41932195
|
29.617
|
|
Sorl1
|
sortilin-related receptor, LDLR class A repeats-containing
|
|
chr11_-_5965544
|
29.563
|
|
Camk2b
|
calcium/calmodulin-dependent protein kinase II, beta
|
|
chr3_-_125641429
|
29.450
|
NM_011674
|
Ugt8a
|
UDP galactosyltransferase 8A
|
|
chr9_-_86775282
|
29.218
|
|
Snap91
|
synaptosomal-associated protein 91
|
|
chr2_+_28048779
|
29.091
|
|
Olfm1
|
olfactomedin 1
|
|
chr9_+_103014374
|
29.090
|
NM_173781
|
Rab6b
|
RAB6B, member RAS oncogene family
|
|
chr5_+_37633353
|
29.085
|
|
Crmp1
|
collapsin response mediator protein 1
|
|
chr5_+_37633317
|
28.736
|
NM_001136058
|
Crmp1
|
collapsin response mediator protein 1
|
|
chr4_+_138011061
|
28.578
|
NM_025451
|
Camk2n1
|
calcium/calmodulin-dependent protein kinase II inhibitor 1
|
|
chr11_-_103815334
|
28.489
|
|
Nsf
|
N-ethylmaleimide sensitive fusion protein
|
|
chr8_+_121971001
|
28.359
|
|
Necab2
|
N-terminal EF-hand calcium binding protein 2
|
|
chr13_-_25361773
|
28.208
|
NM_009513
|
Nrsn1
|
neurensin 1
|
|
chr1_+_60466568
|
27.699
|
|
Abi2
|
abl-interactor 2
|
|
chr4_+_155068604
|
27.347
|
|
B930041F14Rik
|
RIKEN cDNA B930041F14 gene
|
|
chr6_+_22825464
|
26.796
|
NM_001081306
|
Ptprz1
|
protein tyrosine phosphatase, receptor type Z, polypeptide 1
|
|
chr2_-_104250490
|
26.361
|
NM_001033347
|
D430041D05Rik
|
RIKEN cDNA D430041D05 gene
|
|
chr2_+_164793698
|
26.197
|
|
Slc12a5
|
solute carrier family 12, member 5
|
|
chr6_-_23789419
|
26.149
|
|
Cadps2
|
Ca2+-dependent activator protein for secretion 2
|
|
chr15_+_87374667
|
26.116
|
|
Fam19a5
|
family with sequence similarity 19, member A5
|
|
chr4_+_148960536
|
26.090
|
|
Clstn1
|
calsyntenin 1
|
|
chr16_-_4420415
|
25.946
|
|
Adcy9
|
adenylate cyclase 9
|
|
chr2_+_164793487
|
25.792
|
NM_020333
|
Slc12a5
|
solute carrier family 12, member 5
|
|
chr16_+_91270319
|
25.745
|
|
Olig1
|
oligodendrocyte transcription factor 1
|
|
chr5_+_18732863
|
25.457
|
NM_001170746
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2
|
|
chr5_+_27231234
|
24.626
|
NM_001198886
|
Dpp6
|
dipeptidylpeptidase 6
|
|
chr15_+_100701068
|
24.588
|
NM_001077499
|
Scn8a
|
sodium channel, voltage-gated, type VIII, alpha
|
|
chr11_-_5965555
|
24.552
|
|
Camk2b
|
calcium/calmodulin-dependent protein kinase II, beta
|
|
chr5_+_37259780
|
24.505
|
NM_172994
|
Ppp2r2c
|
protein phosphatase 2 (formerly 2A), regulatory subunit B (PR 52), gamma isoform
|
|
chr7_+_138079669
|
24.470
|
NM_019564
|
Htra1
|
HtrA serine peptidase 1
|
|
chr1_+_60466499
|
24.382
|
|
Abi2
|
abl-interactor 2
|
|
chr3_+_68298129
|
24.049
|
|
Schip1
|
schwannomin interacting protein 1
|
|
chr1_+_66221851
|
24.021
|
NM_001039934
NM_008632
|
Mtap2
|
microtubule-associated protein 2
|
|
chr13_-_25029458
|
23.904
|
NM_172532
|
Aldh5a1
|
aldhehyde dehydrogenase family 5, subfamily A1
|
|
chr15_-_75397601
|
23.879
|
NM_011837
|
Ly6h
|
lymphocyte antigen 6 complex, locus H
|
|
chr2_-_130867226
|
23.861
|
|
Gfra4
|
glial cell line derived neurotrophic factor family receptor alpha 4
|
|
chr2_-_130867149
|
23.704
|
|
Gfra4
|
glial cell line derived neurotrophic factor family receptor alpha 4
|
|
chr7_-_17200285
|
23.392
|
NM_172739
|
Grlf1
|
glucocorticoid receptor DNA binding factor 1
|
|
chr2_-_117168706
|
23.379
|
|
Rasgrp1
|
RAS guanyl releasing protein 1
|
|
chrX_+_5624624
|
23.379
|
NM_021431
|
Nudt11
|
nudix (nucleoside diphosphate linked moiety X)-type motif 11
|
|
chr1_+_60466462
|
23.235
|
NM_001198570
NM_001198571
NM_198127
|
Abi2
|
abl-interactor 2
|
|
chr2_-_168567244
|
23.217
|
NM_015731
|
Atp9a
|
ATPase, class II, type 9A
|
|
chr9_+_122481617
|
23.184
|
|
9530059O14Rik
|
RIKEN cDNA 9530059O14 gene
|
|
chr4_+_148960604
|
23.164
|
NM_023051
|
Clstn1
|
calsyntenin 1
|
|
chr2_-_117168409
|
23.003
|
|
Rasgrp1
|
RAS guanyl releasing protein 1
|
|
chr1_-_38954763
|
22.949
|
|
Chst10
|
carbohydrate sulfotransferase 10
|
|
chr14_-_24824081
|
22.928
|
|
Kcnma1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1
|
|
chr5_-_34630913
|
22.887
|
NM_001015039
|
Zfyve28
|
zinc finger, FYVE domain containing 28
|
|
chr16_+_5884885
|
22.740
|
NM_021477
|
Rbfox1
|
RNA binding protein, fox-1 homolog (C. elegans) 1
|
|
chr17_-_24826873
|
22.677
|
NM_011522
|
Syngr3
|
synaptogyrin 3
|
|
chr7_+_133415982
|
22.567
|
NM_145587
|
Sbk1
|
SH3-binding kinase 1
|
|
chr7_+_52960585
|
22.471
|
NM_016974
|
Dbp
|
D site albumin promoter binding protein
|
|
chr11_-_110198978
|
22.402
|
NM_147219
|
Abca5
|
ATP-binding cassette, sub-family A (ABC1), member 5
|
|
chr15_+_78744576
|
22.124
|
|
Pdxp
|
pyridoxal (pyridoxine, vitamin B6) phosphatase
|
|
chr6_-_143048295
|
22.106
|
|
5730419I09Rik
|
RIKEN cDNA 5730419I09 gene
|
|
chr10_+_90292453
|
22.085
|
|
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B
|
|
chr10_-_70061996
|
22.040
|
|
Phyhipl
|
phytanoyl-CoA hydroxylase interacting protein-like
|
|
chr8_+_121970569
|
22.032
|
NM_054095
|
Necab2
|
N-terminal EF-hand calcium binding protein 2
|
|
chr14_-_30534966
|
22.020
|
NM_009785
|
Cacna2d3
|
calcium channel, voltage-dependent, alpha2/delta subunit 3
|
|
chr5_-_122952228
|
21.893
|
NM_001110140
NM_009722
|
Atp2a2
|
ATPase, Ca++ transporting, cardiac muscle, slow twitch 2
|
|
chr1_+_60466620
|
21.875
|
|
Abi2
|
abl-interactor 2
|
|
chr12_-_4038848
|
21.864
|
NM_001082483
|
Efr3b
|
EFR3 homolog B (S. cerevisiae)
|
|
chr9_-_4796141
|
21.689
|
NM_001113180
NM_001113181
NM_019691
|
Gria4
|
glutamate receptor, ionotropic, AMPA4 (alpha 4)
|
|
chr15_-_75397030
|
21.675
|
NM_001135689
|
Ly6h
|
lymphocyte antigen 6 complex, locus H
|
|
chr8_+_11728104
|
21.567
|
NM_001113517
|
Arhgef7
|
Rho guanine nucleotide exchange factor (GEF7)
|
|
chr7_-_88974235
|
21.495
|
NM_146193
|
Btbd1
|
BTB (POZ) domain containing 1
|
|
chr7_+_52960702
|
21.436
|
|
Dbp
|
D site albumin promoter binding protein
|
|
chr17_+_86567003
|
21.224
|
NM_011104
|
Prkce
|
protein kinase C, epsilon
|
|
chr12_-_113146259
|
21.104
|
NM_011625
|
Ppp1r13b
|
protein phosphatase 1, regulatory (inhibitor) subunit 13B
|
|
chr7_+_114514258
|
20.897
|
NM_021889
|
Syt9
|
synaptotagmin IX
|
|
chr6_+_117961386
|
20.685
|
|
Rasgef1a
|
RasGEF domain family, member 1A
|
|
chr6_+_88674398
|
20.628
|
NM_001166249
NM_011844
|
Mgll
|
monoglyceride lipase
|
|
chr5_+_67137054
|
20.602
|
NM_001001980
|
Limch1
|
LIM and calponin homology domains 1
|
|
chr10_-_70062025
|
20.563
|
NM_178621
|
Phyhipl
|
phytanoyl-CoA hydroxylase interacting protein-like
|
|
chr4_+_155068566
|
20.517
|
NM_178699
|
B930041F14Rik
|
RIKEN cDNA B930041F14 gene
|
|
chr5_-_133019167
|
20.383
|
|
Auts2
|
autism susceptibility candidate 2
|
|
chr4_+_155068450
|
20.370
|
|
B930041F14Rik
|
RIKEN cDNA B930041F14 gene
|
|
chr4_+_129662110
|
20.288
|
|
Bai2
|
brain-specific angiogenesis inhibitor 2
|
|
chr9_+_103014445
|
20.239
|
|
Rab6b
|
RAB6B, member RAS oncogene family
|
|
chr6_+_71657791
|
19.880
|
NM_178608
|
Reep1
|
receptor accessory protein 1
|
|
chrX_-_71070284
|
19.780
|
NM_001029868
|
Pdzd4
|
PDZ domain containing 4
|
|
chr19_-_5098498
|
19.473
|
|
Cnih2
|
cornichon homolog 2 (Drosophila)
|
|
chr16_-_20241392
|
19.425
|
NM_198599
|
Map6d1
|
MAP6 domain containing 1
|
|
chr7_+_133416177
|
19.349
|
|
Sbk1
|
SH3-binding kinase 1
|
|
chr2_+_92439758
|
19.296
|
NM_023850
|
Chst1
|
carbohydrate (keratan sulfate Gal-6) sulfotransferase 1
|
|
chr12_+_82732188
|
19.225
|
NM_001033149
|
Ttc9
|
tetratricopeptide repeat domain 9
|
|
chr9_-_21856445
|
19.207
|
NM_010487
|
Elavl3
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 3 (Hu antigen C)
|
|
chr9_+_54547344
|
18.994
|
NM_021422
|
Dnaja4
|
DnaJ (Hsp40) homolog, subfamily A, member 4
|
|
chrX_+_8469507
|
18.935
|
|
B630019K06Rik
|
RIKEN cDNA B630019K06 gene
|
|
chr2_+_90729768
|
18.917
|
|
C1qtnf4
|
C1q and tumor necrosis factor related protein 4
|
|
chr8_-_67373608
|
18.866
|
|
Klhl2
|
kelch-like 2, Mayven (Drosophila)
|
|
chr10_-_115910578
|
18.842
|
NM_021452
|
Kcnmb4
|
potassium large conductance calcium-activated channel, subfamily M, beta member 4
|
|
chr1_-_166388431
|
18.813
|
NM_009721
|
Atp1b1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide
|
|
chr4_+_42930076
|
18.764
|
NM_172690
|
N28178
|
expressed sequence N28178
|
|
chr8_-_89996474
|
18.703
|
NM_019626
|
Cbln1
|
cerebellin 1 precursor protein
|
|
chr2_+_71819264
|
18.593
|
NM_019688
|
Rapgef4
|
Rap guanine nucleotide exchange factor (GEF) 4
|
|
chr11_-_87172419
|
18.554
|
NM_177167
|
Ppm1e
|
protein phosphatase 1E (PP2C domain containing)
|
|
chr2_+_106533281
|
18.493
|
|
Mpped2
|
metallophosphoesterase domain containing 2
|
|
chr2_+_14525932
|
18.476
|
NM_023116
|
Cacnb2
|
calcium channel, voltage-dependent, beta 2 subunit
|
|
chr2_-_118529608
|
18.331
|
NM_001081971
|
Gm1337
|
predicted gene 1337
|
|
chr3_+_34549057
|
18.265
|
|
Sox2
|
SRY-box containing gene 2
|
|
chr1_+_42752608
|
18.204
|
|
|
|
|
chr19_-_5098381
|
18.202
|
NM_009920
|
Cnih2
|
cornichon homolog 2 (Drosophila)
|
|
chr1_+_63492447
|
18.142
|
NM_001177600
NM_011780
|
Adam23
|
a disintegrin and metallopeptidase domain 23
|
|
chr13_+_9275741
|
18.096
|
NM_001081426
|
Dip2c
|
DIP2 disco-interacting protein 2 homolog C (Drosophila)
|
|
chr10_+_19868725
|
18.018
|
NM_001198635
NM_008635
|
Mtap7
|
microtubule-associated protein 7
|
|
chr9_+_20836481
|
17.884
|
NM_008319
|
Icam5
|
intercellular adhesion molecule 5, telencephalin
|
|
chr2_+_156546694
|
17.727
|
NM_001042488
|
Dlgap4
|
discs, large homolog-associated protein 4 (Drosophila)
|
|
chr11_+_86940545
|
17.713
|
NM_197987
|
Trim37
|
tripartite motif-containing 37
|
|
chr15_-_79763138
|
17.555
|
|
Cbx7
|
chromobox homolog 7
|
|
chr13_+_93195850
|
17.508
|
NM_175455
|
Ankrd34b
|
ankyrin repeat domain 34B
|
|
chr9_-_62384483
|
17.408
|
|
Coro2b
|
coronin, actin binding protein, 2B
|
|
chr13_-_69138164
|
17.071
|
|
Adcy2
|
adenylate cyclase 2
|
|
chr2_+_138082301
|
17.067
|
NM_001025431
|
Btbd3
|
BTB (POZ) domain containing 3
|
|
chr18_+_45428413
|
17.057
|
|
Kcnn2
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 2
|
|
chr4_+_129662284
|
16.995
|
NM_173071
|
Bai2
|
brain-specific angiogenesis inhibitor 2
|
|
chr9_+_20836560
|
16.800
|
|
Icam5
|
intercellular adhesion molecule 5, telencephalin
|
|
chr9_-_62384846
|
16.648
|
NM_175484
|
Coro2b
|
coronin, actin binding protein, 2B
|
|
chr13_-_78337844
|
16.645
|
|
Nr2f1
|
nuclear receptor subfamily 2, group F, member 1
|
|
chrX_+_5624820
|
16.637
|
|
Nudt11
|
nudix (nucleoside diphosphate linked moiety X)-type motif 11
|
|
chr13_-_69138229
|
16.442
|
|
Adcy2
|
adenylate cyclase 2
|
|
chr15_+_83610459
|
16.331
|
|
Mpped1
|
metallophosphoesterase domain containing 1
|
|
chr11_+_100436346
|
16.267
|
|
Cnp
|
2',3'-cyclic nucleotide 3' phosphodiesterase
|
|
chr2_+_156547014
|
16.267
|
NM_001042487
|
Dlgap4
|
discs, large homolog-associated protein 4 (Drosophila)
|
|
chr7_+_108469591
|
16.153
|
|
Stard10
|
START domain containing 10
|
|
chr2_-_73613381
|
16.149
|
NM_001113246
NM_001166603
|
Chn1
|
chimerin (chimaerin) 1
|
|
chr2_+_28048511
|
16.079
|
|
Olfm1
|
olfactomedin 1
|
|
chr15_-_83936183
|
15.957
|
NM_013873
|
Sult4a1
|
sulfotransferase family 4A, member 1
|
|
chrX_+_8469478
|
15.822
|
NM_175327
|
B630019K06Rik
|
RIKEN cDNA B630019K06 gene
|
|
chr9_-_69989285
|
15.763
|
NM_029784
|
Fam81a
|
family with sequence similarity 81, member A
|
|
chr11_+_100436228
|
15.619
|
NM_009923
|
Cnp
|
2',3'-cyclic nucleotide 3' phosphodiesterase
|
|
chr14_+_69955194
|
15.592
|
NM_026174
|
Entpd4
|
ectonucleoside triphosphate diphosphohydrolase 4
|
|
chr12_+_83271002
|
15.584
|
NM_001167983
|
Sipa1l1
|
signal-induced proliferation-associated 1 like 1
|
|
chr7_+_53651811
|
15.563
|
NM_008421
NM_001112739
|
Kcnc1
|
potassium voltage gated channel, Shaw-related subfamily, member 1
|
|
chr12_-_11443284
|
15.514
|
NM_012038
|
Vsnl1
|
visinin-like 1
|
|
chr7_-_88599560
|
15.431
|
NM_007755
|
Cpeb1
|
cytoplasmic polyadenylation element binding protein 1
|
|
chr14_-_19726121
|
15.399
|
NM_144839
|
Ube2e2
|
ubiquitin-conjugating enzyme E2E 2 (UBC4/5 homolog, yeast)
|
|
chr6_-_23789048
|
15.274
|
|
Cadps2
|
Ca2+-dependent activator protein for secretion 2
|
|
chr15_-_27955680
|
15.188
|
|
Trio
|
triple functional domain (PTPRF interacting)
|
|
chr9_+_51573434
|
15.104
|
NM_175535
|
Arhgap20
|
Rho GTPase activating protein 20
|
|
chr9_-_69989168
|
15.086
|
|
Fam81a
|
family with sequence similarity 81, member A
|