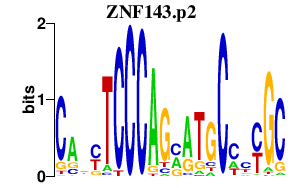
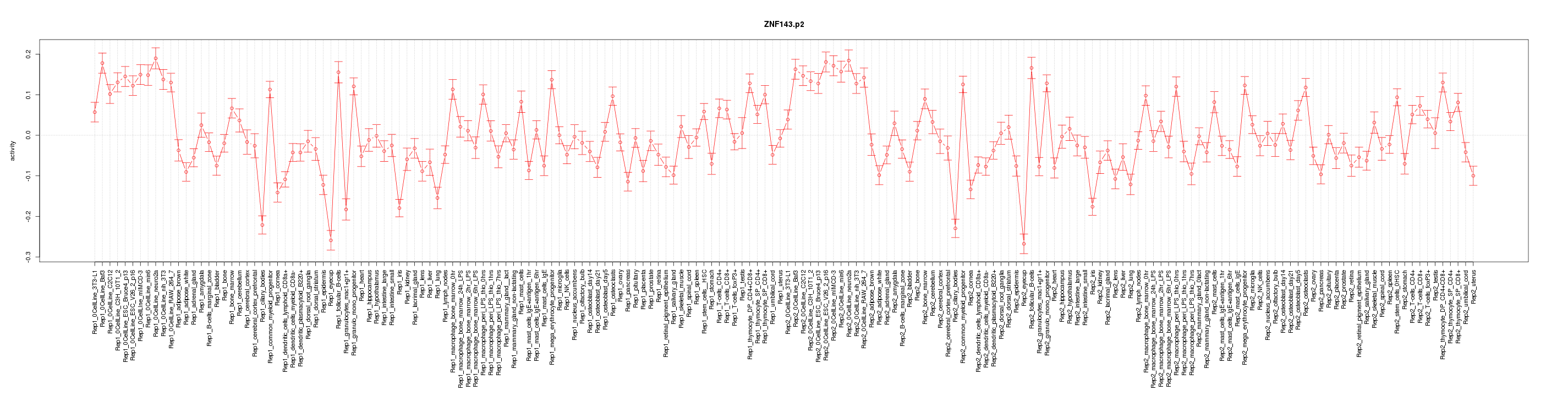
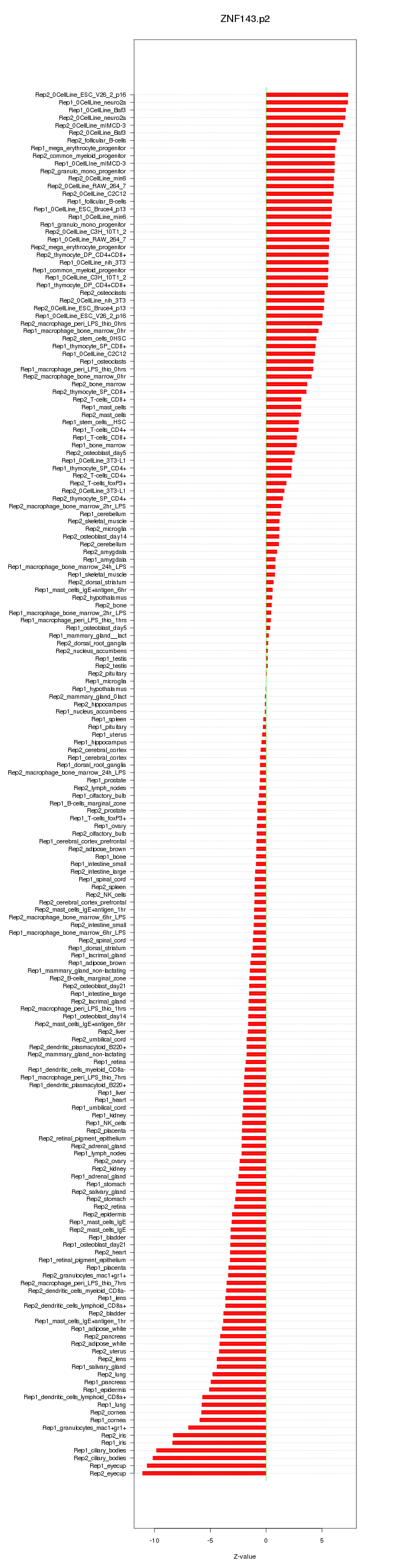
|
chr11_+_117710532
|
70.316
|
NM_001012273
NM_009689
|
Birc5
|
baculoviral IAP repeat-containing 5
|
|
chr5_+_34000745
|
67.111
|
NM_001040435
|
Tacc3
|
transforming, acidic coiled-coil containing protein 3
|
|
chr7_+_13864164
|
62.785
|
NM_010715
|
Lig1
|
ligase I, DNA, ATP-dependent
|
|
chr18_+_34784277
|
44.408
|
NM_001166407
|
Kif20a
|
kinesin family member 20A
|
|
chr10_-_128002934
|
44.133
|
|
Pa2g4
|
proliferation-associated 2G4
|
|
chr18_+_34784589
|
42.417
|
NM_009004
NM_001166406
|
Kif20a
|
kinesin family member 20A
|
|
chr10_-_128003058
|
36.249
|
|
Pa2g4
|
proliferation-associated 2G4
|
|
chr10_-_128002991
|
35.480
|
|
Pa2g4
|
proliferation-associated 2G4
|
|
chr10_-_128002987
|
35.143
|
NM_011119
|
Pa2g4
|
proliferation-associated 2G4
|
|
chr15_-_81560283
|
33.053
|
|
Rangap1
|
RAN GTPase activating protein 1
|
|
chr4_+_107040647
|
31.104
|
|
Tmem48
|
transmembrane protein 48
|
|
chr15_-_81560257
|
31.021
|
|
Rangap1
|
RAN GTPase activating protein 1
|
|
chr15_-_81560302
|
30.436
|
|
Rangap1
|
RAN GTPase activating protein 1
|
|
chr15_-_81560309
|
30.344
|
NM_001146174
NM_011241
|
Rangap1
|
RAN GTPase activating protein 1
|
|
chr8_+_73988823
|
28.436
|
NM_053162
|
Mrpl34
|
mitochondrial ribosomal protein L34
|
|
chr12_-_114067537
|
28.372
|
NM_028023
|
Cdca4
|
cell division cycle associated 4
|
|
chr4_+_107040388
|
27.219
|
NM_028355
|
Tmem48
|
transmembrane protein 48
|
|
chr4_-_86257921
|
27.105
|
NM_173400
|
Haus6
|
HAUS augmin-like complex, subunit 6
|
|
chr9_-_34983552
|
26.249
|
NM_027030
|
Dcps
|
decapping enzyme, scavenger
|
|
chr4_+_107040634
|
26.042
|
|
Tmem48
|
transmembrane protein 48
|
|
chr17_-_23810736
|
22.750
|
NM_001008425
|
Thoc6
|
THO complex 6 homolog (Drosophila)
|
|
chr2_+_118938549
|
21.183
|
NM_011234
|
Rad51
|
RAD51 homolog (S. cerevisiae)
|
|
chr10_-_126748678
|
21.049
|
|
Mars
|
methionine-tRNA synthetase
|
|
chr17_-_23810808
|
20.492
|
|
Thoc6
|
THO complex 6 homolog (Drosophila)
|
|
chr7_+_129302998
|
20.455
|
|
Plk1
|
polo-like kinase 1 (Drosophila)
|
|
chr15_+_102348622
|
20.349
|
NM_009319
|
Tarbp2
|
TAR (HIV) RNA binding protein 2
|
|
chr2_+_119444033
|
20.143
|
NM_001042652
NM_133851
|
Nusap1
|
nucleolar and spindle associated protein 1
|
|
chr10_-_80958977
|
19.946
|
NM_134009
|
Ncln
|
nicalin homolog (zebrafish)
|
|
chr7_+_17323737
|
19.896
|
NM_198613
|
Ap2s1
|
adaptor-related protein complex 2, sigma 1 subunit
|
|
chr7_+_129303029
|
19.762
|
|
Plk1
|
polo-like kinase 1 (Drosophila)
|
|
chr1_-_20810238
|
19.519
|
NM_008563
|
Mcm3
|
minichromosome maintenance deficient 3 (S. cerevisiae)
|
|
chr7_+_129302950
|
19.490
|
NM_011121
|
Plk1
|
polo-like kinase 1 (Drosophila)
|
|
chr8_+_108384479
|
19.206
|
NM_026532
|
Nutf2
|
nuclear transport factor 2
|
|
chr10_+_127452219
|
18.614
|
|
Prim1
|
DNA primase, p49 subunit
|
|
chr19_-_10278231
|
18.417
|
NM_007999
|
Fen1
|
flap structure specific endonuclease 1
|
|
chr7_+_123236814
|
17.757
|
NM_019772
|
1110004F10Rik
|
RIKEN cDNA 1110004F10 gene
|
|
chr3_-_88905775
|
17.476
|
NM_134469
|
Fdps
|
farnesyl diphosphate synthetase
|
|
chr10_+_127452270
|
17.468
|
NM_008921
|
Prim1
|
DNA primase, p49 subunit
|
|
chr19_+_37450854
|
17.397
|
NM_010615
|
Kif11
|
kinesin family member 11
|
|
chr10_-_80380806
|
17.331
|
NM_010722
|
Lmnb2
|
lamin B2
|
|
chr11_+_98769162
|
17.296
|
NM_001025779
|
Cdc6
|
cell division cycle 6 homolog (S. cerevisiae)
|
|
chr11_+_98769122
|
17.150
|
|
Cdc6
|
cell division cycle 6 homolog (S. cerevisiae)
|
|
chr19_+_5601857
|
17.067
|
NM_026616
|
Rnaseh2c
|
ribonuclease H2, subunit C
|
|
chr3_-_96001504
|
16.756
|
NM_026975
|
Bola1
|
bolA-like 1 (E. coli)
|
|
chr2_+_164595389
|
16.732
|
|
Ube2c
|
ubiquitin-conjugating enzyme E2C
|
|
chr2_+_118423946
|
16.718
|
NM_009773
|
Bub1b
|
budding uninhibited by benzimidazoles 1 homolog, beta (S. cerevisiae)
|
|
chr5_+_144165615
|
16.550
|
|
Kdelr2
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 2
|
|
chr9_+_107489981
|
16.504
|
NM_025903
|
Ifrd2
|
interferon-related developmental regulator 2
|
|
chr14_-_55136145
|
16.358
|
NM_013768
|
Prmt5
|
protein arginine N-methyltransferase 5
|
|
chr2_+_118424004
|
16.353
|
|
Bub1b
|
budding uninhibited by benzimidazoles 1 homolog, beta (S. cerevisiae)
|
|
chr15_-_75760134
|
16.018
|
NM_031201
|
Tsta3
|
tissue specific transplantation antigen P35B
|
|
chr2_-_84567398
|
15.906
|
|
Clp1
|
CLP1, cleavage and polyadenylation factor I subunit, homolog (S. cerevisiae)
|
|
chr2_-_84567424
|
15.766
|
NM_133840
|
Clp1
|
CLP1, cleavage and polyadenylation factor I subunit, homolog (S. cerevisiae)
|
|
chr8_+_108734906
|
15.739
|
NM_145404
|
Prmt7
|
protein arginine N-methyltransferase 7
|
|
chr2_+_164595447
|
15.569
|
|
Ube2c
|
ubiquitin-conjugating enzyme E2C
|
|
chr12_+_107248496
|
15.510
|
|
Vrk1
|
vaccinia related kinase 1
|
|
chr2_+_164595415
|
15.419
|
NM_026785
|
Ube2c
|
ubiquitin-conjugating enzyme E2C
|
|
chr15_+_102126735
|
15.313
|
|
Espl1
|
extra spindle poles-like 1 (S. cerevisiae)
|
|
chr19_+_34996847
|
15.274
|
NM_183046
|
Kif20b
|
kinesin family member 20B
|
|
chr15_+_102126686
|
15.273
|
NM_001014976
|
Espl1
|
extra spindle poles-like 1 (S. cerevisiae)
|
|
chr9_+_107490100
|
15.222
|
|
Ifrd2
|
interferon-related developmental regulator 2
|
|
chr11_+_95528269
|
15.068
|
NM_008831
|
Phb
|
prohibitin
|
|
chr17_+_56442759
|
14.878
|
NM_001111079
NM_010931
|
Uhrf1
|
ubiquitin-like, containing PHD and RING finger domains, 1
|
|
chr4_-_124614072
|
14.812
|
NM_026560
|
Cdca8
|
cell division cycle associated 8
|
|
chr5_+_140895245
|
14.633
|
NM_133916
|
Eif3b
|
eukaryotic translation initiation factor 3, subunit B
|
|
chr5_-_34000478
|
14.400
|
NM_026698
|
Tmem129
|
transmembrane protein 129
|
|
chrX_+_7305654
|
14.273
|
NM_138602
|
Praf2
|
PRA1 domain family 2
|
|
chr18_-_24178797
|
13.868
|
|
Zfp191
|
zinc finger protein 191
|
|
chr17_-_85190113
|
13.812
|
NM_028233
|
Lrpprc
|
leucine-rich PPR-motif containing
|
|
chr3_+_68808879
|
13.610
|
NM_133786
|
Smc4
|
structural maintenance of chromosomes 4
|
|
chr4_-_116380924
|
13.602
|
NM_025962
|
Mmachc
|
methylmalonic aciduria cblC type, with homocystinuria
|
|
chr2_-_154900132
|
13.391
|
|
Ahcy
|
S-adenosylhomocysteine hydrolase
|
|
chr17_-_35810624
|
13.292
|
NM_010364
|
Gtf2h4
|
general transcription factor II H, polypeptide 4
|
|
chr16_-_18289209
|
13.290
|
NM_033324
|
Dgcr8
|
DiGeorge syndrome critical region gene 8
|
|
chr5_+_124199713
|
13.181
|
NM_001042421
|
Kntc1
|
kinetochore associated 1
|
|
chr6_-_34126949
|
13.180
|
NM_021435
|
Slc35b4
|
solute carrier family 35, member B4
|
|
chr13_-_58316923
|
13.180
|
|
Ubqln1
|
ubiquilin 1
|
|
chr10_+_74980744
|
13.057
|
NM_026095
|
Snrpd3
|
small nuclear ribonucleoprotein D3
|
|
chr10_+_92916150
|
13.056
|
|
Lta4h
|
leukotriene A4 hydrolase
|
|
chr2_-_154900188
|
13.040
|
NM_016661
|
Ahcy
|
S-adenosylhomocysteine hydrolase
|
|
chr2_+_90519527
|
13.037
|
|
Nup160
|
nucleoporin 160
|
|
chr1_-_44159187
|
13.019
|
NM_029368
|
1700029F09Rik
|
RIKEN cDNA 1700029F09 gene
|
|
chr10_-_126748757
|
12.925
|
NM_001003913
NM_001171582
|
Mars
|
methionine-tRNA synthetase
|
|
chr12_-_80292805
|
12.888
|
NM_021557
|
Rdh11
|
retinol dehydrogenase 11
|
|
chr6_+_85381958
|
12.828
|
NM_144918
|
Smyd5
|
SET and MYND domain containing 5
|
|
chr17_-_85189859
|
12.811
|
|
Lrpprc
|
leucine-rich PPR-motif containing
|
|
chr2_-_154900160
|
12.756
|
|
Ahcy
|
S-adenosylhomocysteine hydrolase
|
|
chr3_-_54539285
|
12.668
|
NM_001163570
NM_027148
|
Exosc8
|
exosome component 8
|
|
chr2_+_172847371
|
12.615
|
NM_019547
|
Rbm38
|
RNA binding motif protein 38
|
|
chr8_-_12672048
|
12.585
|
NM_198031
|
Tubgcp3
|
tubulin, gamma complex associated protein 3
|
|
chr6_-_92133945
|
12.463
|
NM_025578
|
Mrps25
|
mitochondrial ribosomal protein S25
|
|
chr5_-_23929349
|
12.425
|
|
Cdk5
|
cyclin-dependent kinase 5
|
|
chr10_+_74980789
|
12.410
|
|
Snrpd3
|
small nuclear ribonucleoprotein D3
|
|
chrX_-_7182450
|
12.408
|
NM_138603
|
Ccdc22
|
coiled-coil domain containing 22
|
|
chr7_-_30954705
|
12.117
|
NM_011980
|
Zfp146
|
zinc finger protein 146
|
|
chr7_-_52241669
|
12.005
|
NM_019830
|
Prmt1
|
protein arginine N-methyltransferase 1
|
|
chr9_-_103104394
|
11.960
|
NM_009275
|
Srprb
|
signal recognition particle receptor, B subunit
|
|
chr4_-_49533914
|
11.947
|
NM_178603
|
Mrpl50
|
mitochondrial ribosomal protein L50
|
|
chr3_+_79433000
|
11.896
|
NM_029482
|
4930579G24Rik
|
RIKEN cDNA 4930579G24 gene
|
|
chr1_+_193645351
|
11.833
|
NM_010892
|
Nek2
|
NIMA (never in mitosis gene a)-related expressed kinase 2
|
|
chr19_+_6334978
|
11.811
|
NM_001168488
NM_001168489
NM_001168490
NM_008583
|
Men1
|
multiple endocrine neoplasia 1
|
|
chr11_-_60692533
|
11.732
|
NM_001168507
NM_173453
|
Tmem11
|
transmembrane protein 11
|
|
chr4_-_129318200
|
11.712
|
|
Txlna
|
taxilin alpha
|
|
chr8_-_108734817
|
11.667
|
NM_001007567
|
Slc7a6os
|
solute carrier family 7, member 6 opposite strand
|
|
chr9_+_26807281
|
11.665
|
|
Thyn1
|
thymocyte nuclear protein 1
|
|
chr18_-_78006524
|
11.571
|
|
Haus1
|
HAUS augmin-like complex, subunit 1
|
|
chr2_+_128926721
|
11.470
|
NM_009086
|
Polr1b
|
polymerase (RNA) I polypeptide B
|
|
chr19_+_46130978
|
11.409
|
NM_001081214
|
Pprc1
|
peroxisome proliferative activated receptor, gamma, coactivator-related 1
|
|
chr2_-_153067043
|
11.409
|
NM_018807
|
Plagl2
|
pleiomorphic adenoma gene-like 2
|
|
chr4_-_126738303
|
11.364
|
NM_026670
|
Zmym1
|
zinc finger, MYM domain containing 1
|
|
chr5_-_34000546
|
11.335
|
|
Tmem129
|
transmembrane protein 129
|
|
chrX_-_136533089
|
11.275
|
NM_001172147
NM_001172148
NM_153586
|
Rbm41
|
RNA binding motif protein 41
|
|
chr4_-_138908210
|
11.184
|
NM_023536
|
Mrto4
|
MRT4, mRNA turnover 4, homolog (S. cerevisiae)
|
|
chr1_+_136859153
|
11.142
|
NM_026024
|
Ube2t
|
ubiquitin-conjugating enzyme E2T (putative)
|
|
chr8_-_108375786
|
11.114
|
NM_177150
|
Cenpt
|
centromere protein T
|
|
chr2_+_70499545
|
11.106
|
NM_027352
|
Gorasp2
|
golgi reassembly stacking protein 2
|
|
chr15_+_85690136
|
11.027
|
NM_001168672
|
Gtse1
|
G two S phase expressed protein 1
|
|
chr4_+_134066913
|
11.024
|
NM_001081099
|
2610002D18Rik
|
RIKEN cDNA 2610002D18 gene
|
|
chr1_+_136859183
|
10.967
|
|
Ube2t
|
ubiquitin-conjugating enzyme E2T (putative)
|
|
chr15_+_85690375
|
10.940
|
NM_013882
|
Gtse1
|
G two S phase expressed protein 1
|
|
chr9_+_26807258
|
10.933
|
NM_144543
|
Thyn1
|
thymocyte nuclear protein 1
|
|
chr10_-_80363422
|
10.903
|
|
Timm13
|
translocase of inner mitochondrial membrane 13 homolog (yeast)
|
|
chr2_+_153171730
|
10.845
|
NM_001039939
|
Asxl1
|
additional sex combs like 1 (Drosophila)
|
|
chr16_-_87496048
|
10.838
|
NM_009840
|
Cct8
|
chaperonin containing Tcp1, subunit 8 (theta)
|
|
chr14_-_62058627
|
10.776
|
NM_008466
|
Kpna3
|
karyopherin (importin) alpha 3
|
|
chr16_-_16359122
|
10.603
|
NM_001025947
NM_152816
|
Dnm1l
|
dynamin 1-like
|
|
chr8_-_59966523
|
10.567
|
NM_021788
|
Sap30
|
sin3 associated polypeptide
|
|
chr6_-_112646587
|
10.452
|
NM_001167730
NM_021385
|
Rad18
|
RAD18 homolog (S. cerevisiae)
|
|
chr11_+_69393851
|
10.451
|
NM_001127233
NM_011640
|
Trp53
|
transformation related protein 53
|
|
chr15_-_77801154
|
10.415
|
NM_018749
|
Eif3d
|
eukaryotic translation initiation factor 3, subunit D
|
|
chr9_+_48863746
|
10.332
|
|
Zw10
|
ZW10 homolog (Drosophila), centromere/kinetochore protein
|
|
chr11_-_4604308
|
10.320
|
NM_197979
|
Uqcr10
|
ubiquinol-cytochrome c reductase, complex III subunit X
|
|
chr6_+_35127603
|
10.234
|
NM_027513
|
Nup205
|
nucleoporin 205
|
|
chr4_-_120597563
|
10.204
|
NM_001160043
NM_028457
|
Dem1
|
defects in morphology 1 homolog (S. cerevisiae)
|
|
chr13_-_35998160
|
10.121
|
NM_145938
|
Rpp40
|
ribonuclease P 40 subunit (human)
|
|
chr2_+_130100145
|
10.031
|
NM_024193
|
Nop56
|
NOP56 ribonucleoprotein homolog (yeast)
|
|
chr15_-_9070066
|
10.025
|
NM_013787
NM_145468
|
Skp2
|
S-phase kinase-associated protein 2 (p45)
|
|
chr4_-_129317895
|
10.009
|
|
Txlna
|
taxilin alpha
|
|
chr10_-_80363493
|
9.987
|
|
Timm13
|
translocase of inner mitochondrial membrane 13 homolog (yeast)
|
|
chr8_+_96738490
|
9.866
|
NM_172410
|
Nup93
|
nucleoporin 93
|
|
chr1_+_173433608
|
9.863
|
NM_019484
|
Refbp2
|
RNA and export factor binding protein 2
|
|
chr7_-_38892432
|
9.833
|
NM_007633
|
Ccne1
|
cyclin E1
|
|
chr6_-_83004374
|
9.811
|
NM_019752
|
Htra2
|
HtrA serine peptidase 2
|
|
chr11_+_62416376
|
9.788
|
|
2410006H16Rik
|
RIKEN cDNA 2410006H16 gene
|
|
chr11_+_70833337
|
9.780
|
|
Mis12
|
MIS12 homolog (yeast)
|
|
chr8_+_96738448
|
9.774
|
|
Nup93
|
nucleoporin 93
|
|
chr15_+_90054711
|
9.721
|
NM_001033441
|
Alg10b
|
asparagine-linked glycosylation 10 homolog B (yeast, alpha-1,2-glucosyltransferase)
|
|
chr9_+_20833816
|
9.711
|
NM_023892
|
Icam4
|
intercellular adhesion molecule 4, Landsteiner-Wiener blood group
|
|
chr11_-_60692261
|
9.702
|
|
Tmem11
|
transmembrane protein 11
|
|
chr11_+_51781124
|
9.688
|
NM_029976
|
Cdkn2aipnl
|
CDKN2A interacting protein N-terminal like
|
|
chr7_+_73205221
|
9.683
|
NM_021336
|
Snrpa1
|
small nuclear ribonucleoprotein polypeptide A'
|
|
chr16_-_17111300
|
9.651
|
NM_144954
|
Ppil2
|
peptidylprolyl isomerase (cyclophilin)-like 2
|
|
chr10_-_80363606
|
9.549
|
NM_013895
|
Timm13
|
translocase of inner mitochondrial membrane 13 homolog (yeast)
|
|
chr2_-_69044179
|
9.542
|
NM_025565
|
Spc25
|
SPC25, NDC80 kinetochore complex component, homolog (S. cerevisiae)
|
|
chr9_-_57400530
|
9.497
|
NM_025837
|
Mpi
|
mannose phosphate isomerase
|
|
chr12_-_66273564
|
9.397
|
NM_172578
|
Mis18bp1
|
MIS18 binding protein 1
|
|
chr13_-_58316960
|
9.394
|
NM_026842
NM_152234
|
Ubqln1
|
ubiquilin 1
|
|
chr18_-_78006491
|
9.362
|
NM_146089
|
Haus1
|
HAUS augmin-like complex, subunit 1
|
|
chr11_-_106860803
|
9.313
|
NM_010655
|
Kpna2
|
karyopherin (importin) alpha 2
|
|
chr19_-_9973901
|
9.274
|
NM_016692
|
Incenp
|
inner centromere protein
|
|
chr11_-_101413104
|
9.259
|
NM_009764
|
Brca1
|
breast cancer 1
|
|
chr16_+_18498805
|
9.218
|
NM_001081682
|
Gnb1l
|
guanine nucleotide binding protein (G protein), beta polypeptide 1-like
|
|
chr12_-_80292830
|
9.195
|
|
Rdh11
|
retinol dehydrogenase 11
|
|
chr10_-_122633922
|
9.165
|
NM_027604
|
Usp15
|
ubiquitin specific peptidase 15
|
|
chr8_+_113443764
|
9.113
|
NM_009179
|
St3gal2
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 2
|
|
chr16_+_44724394
|
9.105
|
NM_145972
|
BC027231
|
cDNA sequence BC027231
|
|
chr5_+_23900197
|
9.083
|
|
Abcb8
|
ATP-binding cassette, sub-family B (MDR/TAP), member 8
|
|
chr7_+_73205454
|
9.072
|
|
Snrpa1
|
small nuclear ribonucleoprotein polypeptide A'
|
|
chr11_-_115873976
|
8.934
|
NM_016905
|
Galk1
|
galactokinase 1
|
|
chr11_-_107050573
|
8.932
|
NM_001161329
NM_133702
|
Nol11
|
nucleolar protein 11
|
|
chr4_+_62069801
|
8.856
|
NM_027297
|
Prpf4
|
PRP4 pre-mRNA processing factor 4 homolog (yeast)
|
|
chr1_-_179726514
|
8.794
|
NM_007422
|
Adss
|
adenylosuccinate synthetase, non muscle
|
|
chr2_+_166888719
|
8.745
|
|
1500012F01Rik
|
RIKEN cDNA 1500012F01 gene
|
|
chr5_-_24083072
|
8.678
|
|
Abcf2
|
ATP-binding cassette, sub-family F (GCN20), member 2
|
|
chr11_-_106860750
|
8.667
|
|
Kpna2
|
karyopherin (importin) alpha 2
|
|
chr4_-_132399023
|
8.639
|
NM_146154
|
Ppp1r8
|
protein phosphatase 1, regulatory (inhibitor) subunit 8
|
|
chr18_-_85120825
|
8.637
|
|
1700034H14Rik
|
RIKEN cDNA 1700034H14 gene
|
|
chr7_+_73205419
|
8.634
|
|
Snrpa1
|
small nuclear ribonucleoprotein polypeptide A'
|
|
chr12_-_114067236
|
8.567
|
|
Cdca4
|
cell division cycle associated 4
|
|
chr8_-_110112368
|
8.561
|
NM_010817
|
Psmd7
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 7
|
|
chr2_+_152673699
|
8.502
|
NM_001141975
NM_001141976
NM_001141977
NM_001141978
NM_028109
|
Tpx2
|
TPX2, microtubule-associated protein homolog (Xenopus laevis)
|
|
chr12_+_85291557
|
8.490
|
NM_023633
|
2410016O06Rik
|
RIKEN cDNA 2410016O06 gene
|
|
chr17_+_26389911
|
8.490
|
|
Luc7l
|
Luc7 homolog (S. cerevisiae)-like
|
|
chr11_-_70783381
|
8.425
|
|
Nup88
|
nucleoporin 88
|
|
chr2_+_119444452
|
8.425
|
|
Nusap1
|
nucleolar and spindle associated protein 1
|
|
chr11_-_70783455
|
8.419
|
NM_001083331
NM_172394
|
Nup88
|
nucleoporin 88
|
|
chr7_+_51723446
|
8.405
|
|
Josd2
|
Josephin domain containing 2
|
|
chr5_-_145952452
|
8.401
|
NM_020582
|
Atp5j2
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit F2
|
|
chr7_+_108812146
|
8.393
|
NM_009191
|
Clpb
|
ClpB caseinolytic peptidase B homolog (E. coli)
|
|
chr10_-_117147068
|
8.390
|
|
Mdm2
|
transformed mouse 3T3 cell double minute 2
|
|
chr7_+_150255257
|
8.307
|
NM_001115085
NM_020285
NM_138631
|
Tssc4
|
tumor-suppressing subchromosomal transferable fragment 4
|
|
chr11_+_70783966
|
8.212
|
|
Rpain
|
RPA interacting protein
|
|
chr1_+_192886883
|
8.200
|
NM_198654
|
Nsl1
|
NSL1, MIND kinetochore complex component, homolog (S. cerevisiae)
|
|
chr9_+_13553629
|
8.181
|
|
Mtmr2
|
myotubularin related protein 2
|
|
chr5_-_24083185
|
8.180
|
NM_001190443
NM_013853
|
Abcf2
|
ATP-binding cassette, sub-family F (GCN20), member 2
|
|
chr5_+_34001199
|
8.165
|
|
Tacc3
|
transforming, acidic coiled-coil containing protein 3
|
|
chr3_-_104621135
|
8.160
|
NM_001163441
NM_008619
|
Mov10
|
Moloney leukemia virus 10
|
|
chr17_-_85189893
|
8.150
|
|
Lrpprc
|
leucine-rich PPR-motif containing
|
|
chr1_-_181733742
|
8.057
|
|
Ahctf1
|
AT hook containing transcription factor 1
|
|
chr4_-_116855158
|
8.039
|
NM_134471
|
Kif2c
|
kinesin family member 2C
|