Project
Illumina Body Map 2, young vs old
Navigation
Downloads
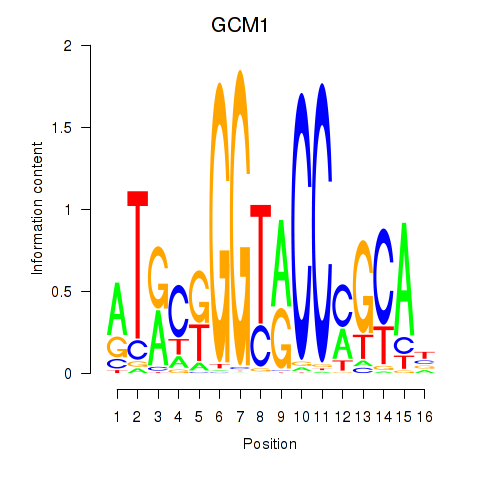
Results for GCM1
Z-value: 2.17
Transcription factors associated with GCM1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
GCM1
|
ENSG00000137270.10 | glial cells missing transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| GCM1 | hg19_v2_chr6_-_53013620_53013644 | 0.06 | 7.4e-01 | Click! |
Activity profile of GCM1 motif
Sorted Z-values of GCM1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_89630186 | 5.22 |
ENST00000390264.2
|
IGKV2-40
|
immunoglobulin kappa variable 2-40 |
| chr22_+_22764088 | 5.14 |
ENST00000390299.2
|
IGLV1-40
|
immunoglobulin lambda variable 1-40 |
| chr22_+_22786288 | 5.13 |
ENST00000390301.2
|
IGLV1-36
|
immunoglobulin lambda variable 1-36 |
| chr22_+_22712087 | 5.11 |
ENST00000390294.2
|
IGLV1-47
|
immunoglobulin lambda variable 1-47 |
| chr2_-_89385283 | 4.84 |
ENST00000390252.2
|
IGKV3-15
|
immunoglobulin kappa variable 3-15 |
| chr22_+_23154239 | 4.55 |
ENST00000390315.2
|
IGLV3-10
|
immunoglobulin lambda variable 3-10 |
| chr22_+_22676808 | 4.51 |
ENST00000390290.2
|
IGLV1-51
|
immunoglobulin lambda variable 1-51 |
| chr2_+_208527094 | 4.46 |
ENST00000429730.1
|
AC079767.4
|
AC079767.4 |
| chr22_+_23165153 | 4.20 |
ENST00000390317.2
|
IGLV2-8
|
immunoglobulin lambda variable 2-8 |
| chr22_+_23077065 | 3.79 |
ENST00000390310.2
|
IGLV2-18
|
immunoglobulin lambda variable 2-18 |
| chr22_+_22930626 | 3.78 |
ENST00000390302.2
|
IGLV2-33
|
immunoglobulin lambda variable 2-33 (non-functional) |
| chr22_+_23040274 | 3.66 |
ENST00000390306.2
|
IGLV2-23
|
immunoglobulin lambda variable 2-23 |
| chr7_+_142028105 | 3.59 |
ENST00000390353.2
|
TRBV6-1
|
T cell receptor beta variable 6-1 |
| chr22_+_23101182 | 3.37 |
ENST00000390312.2
|
IGLV2-14
|
immunoglobulin lambda variable 2-14 |
| chr17_-_3599327 | 3.29 |
ENST00000551178.1
ENST00000552276.1 ENST00000547178.1 |
P2RX5
|
purinergic receptor P2X, ligand-gated ion channel, 5 |
| chr17_-_3599492 | 3.12 |
ENST00000435558.1
ENST00000345901.3 |
P2RX5
|
purinergic receptor P2X, ligand-gated ion channel, 5 |
| chr16_+_3115378 | 3.02 |
ENST00000529550.1
ENST00000551122.1 ENST00000525643.2 ENST00000548807.1 ENST00000528163.2 |
IL32
|
interleukin 32 |
| chr16_+_3115611 | 2.92 |
ENST00000530890.1
ENST00000444393.3 ENST00000533097.2 ENST00000008180.9 ENST00000396890.2 ENST00000525228.1 ENST00000548652.1 ENST00000525377.2 ENST00000530538.2 ENST00000549213.1 ENST00000552936.1 ENST00000548476.1 ENST00000552664.1 ENST00000552356.1 ENST00000551513.1 ENST00000382213.3 ENST00000548246.1 |
IL32
|
interleukin 32 |
| chr22_+_37318624 | 2.77 |
ENST00000421539.1
|
CSF2RB
|
colony stimulating factor 2 receptor, beta, low-affinity (granulocyte-macrophage) |
| chr2_-_136875712 | 2.63 |
ENST00000241393.3
|
CXCR4
|
chemokine (C-X-C motif) receptor 4 |
| chr7_-_142131914 | 2.60 |
ENST00000390375.2
|
TRBV5-6
|
T cell receptor beta variable 5-6 |
| chr9_-_35650900 | 2.44 |
ENST00000259608.3
|
SIT1
|
signaling threshold regulating transmembrane adaptor 1 |
| chr6_-_41122063 | 2.44 |
ENST00000426005.2
ENST00000437044.2 ENST00000373127.4 |
TREML1
|
triggering receptor expressed on myeloid cells-like 1 |
| chr3_+_10206545 | 2.37 |
ENST00000256458.4
|
IRAK2
|
interleukin-1 receptor-associated kinase 2 |
| chr22_+_22730353 | 2.37 |
ENST00000390296.2
|
IGLV5-45
|
immunoglobulin lambda variable 5-45 |
| chr14_+_22180536 | 2.33 |
ENST00000390424.2
|
TRAV2
|
T cell receptor alpha variable 2 |
| chr17_-_73839792 | 2.31 |
ENST00000590762.1
|
UNC13D
|
unc-13 homolog D (C. elegans) |
| chr2_-_89459813 | 2.24 |
ENST00000390256.2
|
IGKV6-21
|
immunoglobulin kappa variable 6-21 (non-functional) |
| chr7_-_38339890 | 2.20 |
ENST00000390341.2
|
TRGV10
|
T cell receptor gamma variable 10 (non-functional) |
| chr22_+_22781853 | 2.19 |
ENST00000390300.2
|
IGLV5-37
|
immunoglobulin lambda variable 5-37 |
| chr1_-_27953031 | 2.19 |
ENST00000374003.3
|
FGR
|
feline Gardner-Rasheed sarcoma viral oncogene homolog |
| chr5_-_55224569 | 2.14 |
ENST00000595799.1
|
AC008914.1
|
Uncharacterized protein |
| chr12_+_8666126 | 2.12 |
ENST00000299665.2
|
CLEC4D
|
C-type lectin domain family 4, member D |
| chr12_-_122238464 | 2.10 |
ENST00000546227.1
|
RHOF
|
ras homolog family member F (in filopodia) |
| chr22_+_37318082 | 2.04 |
ENST00000406230.1
|
CSF2RB
|
colony stimulating factor 2 receptor, beta, low-affinity (granulocyte-macrophage) |
| chr2_+_90060377 | 1.88 |
ENST00000436451.2
|
IGKV6D-21
|
immunoglobulin kappa variable 6D-21 (non-functional) |
| chr11_-_64511789 | 1.87 |
ENST00000419843.1
ENST00000394430.1 |
RASGRP2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
| chrX_-_21676442 | 1.83 |
ENST00000379499.2
|
KLHL34
|
kelch-like family member 34 |
| chr2_+_90108504 | 1.80 |
ENST00000390271.2
|
IGKV6D-41
|
immunoglobulin kappa variable 6D-41 (non-functional) |
| chr4_-_57547870 | 1.78 |
ENST00000381260.3
ENST00000420433.1 ENST00000554144.1 ENST00000557328.1 |
HOPX
|
HOP homeobox |
| chr4_-_57547454 | 1.65 |
ENST00000556376.2
|
HOPX
|
HOP homeobox |
| chr6_-_41715128 | 1.62 |
ENST00000356667.4
ENST00000373025.3 ENST00000425343.2 |
PGC
|
progastricsin (pepsinogen C) |
| chr22_+_22936998 | 1.59 |
ENST00000390303.2
|
IGLV3-32
|
immunoglobulin lambda variable 3-32 (non-functional) |
| chr3_+_130150307 | 1.52 |
ENST00000512836.1
|
COL6A5
|
collagen, type VI, alpha 5 |
| chr11_-_107729287 | 1.49 |
ENST00000375682.4
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr20_+_30639991 | 1.45 |
ENST00000534862.1
ENST00000538448.1 ENST00000375862.2 |
HCK
|
hemopoietic cell kinase |
| chr9_-_136004782 | 1.44 |
ENST00000393157.3
|
RALGDS
|
ral guanine nucleotide dissociation stimulator |
| chr7_-_38331679 | 1.40 |
ENST00000390340.2
|
TRGV11
|
T cell receptor gamma variable 11 (non-functional) |
| chr11_-_64511575 | 1.40 |
ENST00000431822.1
ENST00000377486.3 ENST00000394432.3 |
RASGRP2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
| chr19_-_7797045 | 1.38 |
ENST00000328853.5
|
CLEC4G
|
C-type lectin domain family 4, member G |
| chr20_+_30225682 | 1.31 |
ENST00000376075.3
|
COX4I2
|
cytochrome c oxidase subunit IV isoform 2 (lung) |
| chr3_+_32148106 | 1.27 |
ENST00000425459.1
ENST00000431009.1 |
GPD1L
|
glycerol-3-phosphate dehydrogenase 1-like |
| chr16_+_10971037 | 1.26 |
ENST00000324288.8
ENST00000381835.5 |
CIITA
|
class II, major histocompatibility complex, transactivator |
| chr1_+_67773044 | 1.23 |
ENST00000262345.1
ENST00000371000.1 |
IL12RB2
|
interleukin 12 receptor, beta 2 |
| chr19_-_47551836 | 1.23 |
ENST00000253047.6
|
TMEM160
|
transmembrane protein 160 |
| chr8_+_142138799 | 1.21 |
ENST00000518668.1
|
DENND3
|
DENN/MADD domain containing 3 |
| chr17_+_15943505 | 1.20 |
ENST00000442828.1
|
AC002553.1
|
Uncharacterized protein |
| chr19_+_2270283 | 1.19 |
ENST00000588673.2
|
OAZ1
|
ornithine decarboxylase antizyme 1 |
| chr4_-_8073705 | 1.19 |
ENST00000514025.1
|
ABLIM2
|
actin binding LIM protein family, member 2 |
| chr14_-_105531759 | 1.18 |
ENST00000329797.3
ENST00000539291.2 ENST00000392585.2 |
GPR132
|
G protein-coupled receptor 132 |
| chr7_-_142251148 | 1.17 |
ENST00000390360.3
|
TRBV6-4
|
T cell receptor beta variable 6-4 |
| chr16_-_74734742 | 1.15 |
ENST00000308807.7
ENST00000573267.1 |
MLKL
|
mixed lineage kinase domain-like |
| chr16_+_67062996 | 1.13 |
ENST00000561924.2
|
CBFB
|
core-binding factor, beta subunit |
| chr12_+_93115281 | 1.12 |
ENST00000549856.1
|
PLEKHG7
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 7 |
| chr15_-_60884706 | 1.11 |
ENST00000449337.2
|
RORA
|
RAR-related orphan receptor A |
| chr3_+_32147997 | 1.09 |
ENST00000282541.5
|
GPD1L
|
glycerol-3-phosphate dehydrogenase 1-like |
| chr17_+_7982800 | 1.09 |
ENST00000399413.3
|
AC129492.6
|
AC129492.6 |
| chr12_+_42624050 | 1.07 |
ENST00000601185.1
|
AC020629.1
|
Uncharacterized protein |
| chr11_-_47470682 | 1.05 |
ENST00000529341.1
ENST00000352508.3 |
RAPSN
|
receptor-associated protein of the synapse |
| chr10_-_105677427 | 1.04 |
ENST00000369764.1
|
OBFC1
|
oligonucleotide/oligosaccharide-binding fold containing 1 |
| chr8_-_57232656 | 1.03 |
ENST00000396721.2
|
SDR16C5
|
short chain dehydrogenase/reductase family 16C, member 5 |
| chr5_-_169725231 | 1.02 |
ENST00000046794.5
|
LCP2
|
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa) |
| chr7_+_129074266 | 1.02 |
ENST00000249344.2
ENST00000435494.2 |
STRIP2
|
striatin interacting protein 2 |
| chr11_+_35198118 | 1.01 |
ENST00000525211.1
ENST00000526000.1 ENST00000279452.6 ENST00000527889.1 |
CD44
|
CD44 molecule (Indian blood group) |
| chr17_+_19122674 | 0.95 |
ENST00000436914.1
|
AC106017.1
|
Uncharacterized protein |
| chr2_+_111878483 | 0.94 |
ENST00000308659.8
ENST00000357757.2 ENST00000393253.2 ENST00000337565.5 ENST00000393256.3 |
BCL2L11
|
BCL2-like 11 (apoptosis facilitator) |
| chr19_-_11494975 | 0.92 |
ENST00000222139.6
ENST00000592375.2 |
EPOR
|
erythropoietin receptor |
| chr11_-_47470591 | 0.91 |
ENST00000524487.1
|
RAPSN
|
receptor-associated protein of the synapse |
| chr20_+_36946029 | 0.91 |
ENST00000417318.1
|
BPI
|
bactericidal/permeability-increasing protein |
| chr17_-_5522731 | 0.90 |
ENST00000576905.1
|
NLRP1
|
NLR family, pyrin domain containing 1 |
| chr4_-_8073554 | 0.89 |
ENST00000510277.1
|
ABLIM2
|
actin binding LIM protein family, member 2 |
| chr1_+_161185032 | 0.88 |
ENST00000367992.3
ENST00000289902.1 |
FCER1G
|
Fc fragment of IgE, high affinity I, receptor for; gamma polypeptide |
| chr3_-_69590486 | 0.88 |
ENST00000497880.1
|
FRMD4B
|
FERM domain containing 4B |
| chr14_+_23006547 | 0.88 |
ENST00000390530.1
|
TRAJ7
|
T cell receptor alpha joining 7 |
| chr1_+_17634689 | 0.87 |
ENST00000375453.1
ENST00000375448.4 |
PADI4
|
peptidyl arginine deiminase, type IV |
| chr16_+_67063036 | 0.86 |
ENST00000290858.6
ENST00000564034.1 |
CBFB
|
core-binding factor, beta subunit |
| chr10_-_43762329 | 0.86 |
ENST00000395810.1
|
RASGEF1A
|
RasGEF domain family, member 1A |
| chr11_+_13690200 | 0.85 |
ENST00000354817.3
|
FAR1
|
fatty acyl CoA reductase 1 |
| chr15_+_83776324 | 0.84 |
ENST00000379390.6
ENST00000379386.4 ENST00000565774.1 ENST00000565982.1 |
TM6SF1
|
transmembrane 6 superfamily member 1 |
| chr2_+_102953608 | 0.84 |
ENST00000311734.2
ENST00000409584.1 |
IL1RL1
|
interleukin 1 receptor-like 1 |
| chr5_+_150157860 | 0.83 |
ENST00000600109.1
|
AC010441.1
|
AC010441.1 |
| chr4_+_76649753 | 0.82 |
ENST00000603759.1
|
USO1
|
USO1 vesicle transport factor |
| chr1_+_67673297 | 0.81 |
ENST00000425614.1
ENST00000395227.1 |
IL23R
|
interleukin 23 receptor |
| chr19_+_12902289 | 0.81 |
ENST00000302754.4
|
JUNB
|
jun B proto-oncogene |
| chr11_-_47470703 | 0.80 |
ENST00000298854.2
|
RAPSN
|
receptor-associated protein of the synapse |
| chr18_+_21269556 | 0.80 |
ENST00000399516.3
|
LAMA3
|
laminin, alpha 3 |
| chr19_-_10491130 | 0.79 |
ENST00000530829.1
ENST00000529370.1 |
TYK2
|
tyrosine kinase 2 |
| chr16_+_67063142 | 0.78 |
ENST00000412916.2
|
CBFB
|
core-binding factor, beta subunit |
| chr22_+_46476192 | 0.78 |
ENST00000443490.1
|
FLJ27365
|
hsa-mir-4763 |
| chr10_-_105677886 | 0.77 |
ENST00000224950.3
|
OBFC1
|
oligonucleotide/oligosaccharide-binding fold containing 1 |
| chr14_-_23285069 | 0.77 |
ENST00000554758.1
ENST00000397528.4 |
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr1_+_44399466 | 0.77 |
ENST00000498139.2
ENST00000491846.1 |
ARTN
|
artemin |
| chr21_+_29911640 | 0.77 |
ENST00000412526.1
ENST00000455939.1 |
LINC00161
|
long intergenic non-protein coding RNA 161 |
| chr16_+_215965 | 0.75 |
ENST00000356815.3
|
HBM
|
hemoglobin, mu |
| chr16_+_67063262 | 0.75 |
ENST00000565389.1
|
CBFB
|
core-binding factor, beta subunit |
| chr12_+_106696581 | 0.75 |
ENST00000547153.1
ENST00000299045.3 ENST00000546625.1 ENST00000553098.1 |
TCP11L2
|
t-complex 11, testis-specific-like 2 |
| chr1_+_158975744 | 0.75 |
ENST00000426592.2
|
IFI16
|
interferon, gamma-inducible protein 16 |
| chr4_-_17783135 | 0.75 |
ENST00000265018.3
|
FAM184B
|
family with sequence similarity 184, member B |
| chr2_-_24583168 | 0.73 |
ENST00000361999.3
|
ITSN2
|
intersectin 2 |
| chr16_-_74734672 | 0.72 |
ENST00000306247.7
ENST00000575686.1 |
MLKL
|
mixed lineage kinase domain-like |
| chr12_-_57824561 | 0.72 |
ENST00000448732.1
|
R3HDM2
|
R3H domain containing 2 |
| chr12_-_6665200 | 0.72 |
ENST00000336604.4
ENST00000396840.2 ENST00000356896.4 |
IFFO1
|
intermediate filament family orphan 1 |
| chr5_+_150157444 | 0.69 |
ENST00000526627.1
|
SMIM3
|
small integral membrane protein 3 |
| chr22_-_50964849 | 0.68 |
ENST00000543927.1
ENST00000423348.1 |
SCO2
|
SCO2 cytochrome c oxidase assembly protein |
| chr12_-_122238913 | 0.68 |
ENST00000537157.1
|
AC084018.1
|
AC084018.1 |
| chr19_+_1248547 | 0.67 |
ENST00000586757.1
ENST00000300952.2 |
MIDN
|
midnolin |
| chr6_+_33589161 | 0.67 |
ENST00000605930.1
|
ITPR3
|
inositol 1,4,5-trisphosphate receptor, type 3 |
| chr5_+_176784837 | 0.65 |
ENST00000408923.3
|
RGS14
|
regulator of G-protein signaling 14 |
| chr7_-_74867509 | 0.64 |
ENST00000426327.3
|
GATSL2
|
GATS protein-like 2 |
| chr11_+_73360024 | 0.64 |
ENST00000540431.1
|
PLEKHB1
|
pleckstrin homology domain containing, family B (evectins) member 1 |
| chr8_+_141521386 | 0.63 |
ENST00000220913.5
ENST00000519533.1 |
CHRAC1
|
chromatin accessibility complex 1 |
| chr7_-_99869799 | 0.63 |
ENST00000436886.2
|
GATS
|
GATS, stromal antigen 3 opposite strand |
| chr5_+_135385202 | 0.62 |
ENST00000514554.1
|
TGFBI
|
transforming growth factor, beta-induced, 68kDa |
| chr7_-_44179972 | 0.62 |
ENST00000446581.1
|
MYL7
|
myosin, light chain 7, regulatory |
| chr18_+_43409037 | 0.62 |
ENST00000546268.1
|
SIGLEC15
|
sialic acid binding Ig-like lectin 15 |
| chr19_+_5690207 | 0.62 |
ENST00000347512.3
|
RPL36
|
ribosomal protein L36 |
| chr6_+_144606817 | 0.61 |
ENST00000433557.1
|
UTRN
|
utrophin |
| chr16_+_30212050 | 0.61 |
ENST00000563322.1
|
SULT1A3
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 3 |
| chr1_-_11907829 | 0.60 |
ENST00000376480.3
|
NPPA
|
natriuretic peptide A |
| chr20_-_3996036 | 0.59 |
ENST00000336095.6
|
RNF24
|
ring finger protein 24 |
| chr16_-_425205 | 0.59 |
ENST00000448854.1
|
TMEM8A
|
transmembrane protein 8A |
| chr16_-_11485922 | 0.59 |
ENST00000599216.1
|
CTD-3088G3.8
|
Protein LOC388210 |
| chr4_+_15704679 | 0.59 |
ENST00000382346.3
|
BST1
|
bone marrow stromal cell antigen 1 |
| chr16_+_81478775 | 0.58 |
ENST00000537098.3
|
CMIP
|
c-Maf inducing protein |
| chr1_+_11796126 | 0.57 |
ENST00000376637.3
|
AGTRAP
|
angiotensin II receptor-associated protein |
| chr11_+_13690249 | 0.57 |
ENST00000532701.1
|
FAR1
|
fatty acyl CoA reductase 1 |
| chr16_-_11617444 | 0.55 |
ENST00000598234.1
|
CTD-3088G3.8
|
Protein LOC388210 |
| chr9_-_115653176 | 0.55 |
ENST00000374228.4
|
SLC46A2
|
solute carrier family 46, member 2 |
| chr7_-_26904317 | 0.55 |
ENST00000345317.2
|
SKAP2
|
src kinase associated phosphoprotein 2 |
| chr1_+_11796177 | 0.55 |
ENST00000400895.2
ENST00000376629.4 ENST00000376627.2 ENST00000314340.5 ENST00000452018.2 ENST00000510878.1 |
AGTRAP
|
angiotensin II receptor-associated protein |
| chr8_+_141522017 | 0.53 |
ENST00000518971.1
ENST00000519618.1 |
CHRAC1
|
chromatin accessibility complex 1 |
| chr18_-_46895066 | 0.52 |
ENST00000583225.1
ENST00000584983.1 ENST00000583280.1 ENST00000581738.1 |
DYM
|
dymeclin |
| chr8_-_144512576 | 0.51 |
ENST00000333480.2
|
MAFA
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog A |
| chr19_+_54705025 | 0.51 |
ENST00000441429.1
|
RPS9
|
ribosomal protein S9 |
| chr19_+_54704718 | 0.51 |
ENST00000391752.1
ENST00000402367.1 ENST00000391751.3 |
RPS9
|
ribosomal protein S9 |
| chr6_+_30029008 | 0.51 |
ENST00000332435.5
ENST00000376782.2 ENST00000359374.4 ENST00000376785.2 |
ZNRD1
|
zinc ribbon domain containing 1 |
| chr19_+_18544045 | 0.50 |
ENST00000599699.2
|
SSBP4
|
single stranded DNA binding protein 4 |
| chr2_+_54951679 | 0.50 |
ENST00000356458.6
|
EML6
|
echinoderm microtubule associated protein like 6 |
| chr20_+_32581738 | 0.50 |
ENST00000333552.5
|
RALY
|
RALY heterogeneous nuclear ribonucleoprotein |
| chrX_+_153146127 | 0.49 |
ENST00000452593.1
ENST00000357566.1 |
LCA10
|
Putative lung carcinoma-associated protein 10 |
| chr19_+_54704990 | 0.49 |
ENST00000391753.2
|
RPS9
|
ribosomal protein S9 |
| chr9_+_4985016 | 0.49 |
ENST00000539801.1
|
JAK2
|
Janus kinase 2 |
| chr19_-_38806560 | 0.48 |
ENST00000591755.1
ENST00000337679.8 ENST00000339413.6 |
YIF1B
|
Yip1 interacting factor homolog B (S. cerevisiae) |
| chr10_+_104678032 | 0.48 |
ENST00000369878.4
ENST00000369875.3 |
CNNM2
|
cyclin M2 |
| chr22_+_19705928 | 0.47 |
ENST00000383045.3
ENST00000438754.2 |
SEPT5
|
septin 5 |
| chr10_+_73975742 | 0.47 |
ENST00000299381.4
|
ANAPC16
|
anaphase promoting complex subunit 16 |
| chr18_+_21529811 | 0.47 |
ENST00000588004.1
|
LAMA3
|
laminin, alpha 3 |
| chr4_+_39640787 | 0.46 |
ENST00000532680.1
|
RP11-539G18.2
|
RP11-539G18.2 |
| chr19_-_41945804 | 0.46 |
ENST00000221943.9
ENST00000597457.1 ENST00000589970.1 ENST00000595425.1 ENST00000438807.3 ENST00000589102.1 ENST00000592922.2 |
ATP5SL
|
ATP5S-like |
| chr19_+_50936142 | 0.45 |
ENST00000357701.5
|
MYBPC2
|
myosin binding protein C, fast type |
| chrX_-_27417088 | 0.45 |
ENST00000608735.1
ENST00000422048.1 |
RP11-268G12.1
|
RP11-268G12.1 |
| chr7_+_99971068 | 0.45 |
ENST00000198536.2
ENST00000453419.1 |
PILRA
|
paired immunoglobin-like type 2 receptor alpha |
| chr5_-_142780280 | 0.45 |
ENST00000424646.2
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr14_+_65878650 | 0.44 |
ENST00000555559.1
|
FUT8
|
fucosyltransferase 8 (alpha (1,6) fucosyltransferase) |
| chr16_+_2285817 | 0.44 |
ENST00000564065.1
|
DNASE1L2
|
deoxyribonuclease I-like 2 |
| chr19_+_53510765 | 0.44 |
ENST00000596769.1
|
CTD-2620I22.3
|
CTD-2620I22.3 |
| chr12_-_4554780 | 0.44 |
ENST00000228837.2
|
FGF6
|
fibroblast growth factor 6 |
| chr3_-_46506358 | 0.43 |
ENST00000417439.1
ENST00000431944.1 |
LTF
|
lactotransferrin |
| chr3_-_46506563 | 0.43 |
ENST00000231751.4
|
LTF
|
lactotransferrin |
| chr17_-_39140549 | 0.43 |
ENST00000377755.4
|
KRT40
|
keratin 40 |
| chr16_-_833370 | 0.43 |
ENST00000442466.1
|
MSLNL
|
mesothelin-like |
| chr16_+_29973351 | 0.43 |
ENST00000602948.1
ENST00000279396.6 ENST00000575829.2 ENST00000561899.2 |
TMEM219
|
transmembrane protein 219 |
| chr19_-_38806390 | 0.42 |
ENST00000589247.1
ENST00000329420.8 ENST00000591784.1 |
YIF1B
|
Yip1 interacting factor homolog B (S. cerevisiae) |
| chr20_-_1306351 | 0.41 |
ENST00000381812.1
|
SDCBP2
|
syndecan binding protein (syntenin) 2 |
| chr19_-_6379069 | 0.41 |
ENST00000597721.1
|
PSPN
|
persephin |
| chr17_-_7218631 | 0.40 |
ENST00000577040.2
ENST00000389167.5 ENST00000391950.3 |
GPS2
|
G protein pathway suppressor 2 |
| chr11_-_8986474 | 0.40 |
ENST00000525069.1
|
TMEM9B
|
TMEM9 domain family, member B |
| chr14_+_29299437 | 0.39 |
ENST00000550827.1
ENST00000548087.1 ENST00000551588.1 ENST00000550122.1 |
CTD-2384A14.1
|
CTD-2384A14.1 |
| chr7_-_116607526 | 0.39 |
ENST00000420664.1
|
AC106873.4
|
Uncharacterized protein |
| chr3_-_9994021 | 0.39 |
ENST00000411976.2
ENST00000412055.1 |
PRRT3
|
proline-rich transmembrane protein 3 |
| chr10_-_31146615 | 0.39 |
ENST00000444692.2
|
ZNF438
|
zinc finger protein 438 |
| chr19_-_38806540 | 0.39 |
ENST00000592694.1
|
YIF1B
|
Yip1 interacting factor homolog B (S. cerevisiae) |
| chr3_+_46448648 | 0.39 |
ENST00000399036.3
|
CCRL2
|
chemokine (C-C motif) receptor-like 2 |
| chr11_-_8986279 | 0.38 |
ENST00000534025.1
|
TMEM9B
|
TMEM9 domain family, member B |
| chr17_+_40913210 | 0.38 |
ENST00000253796.5
|
RAMP2
|
receptor (G protein-coupled) activity modifying protein 2 |
| chr2_+_149402009 | 0.38 |
ENST00000457184.1
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chr18_+_21269404 | 0.37 |
ENST00000313654.9
|
LAMA3
|
laminin, alpha 3 |
| chr6_+_42018251 | 0.37 |
ENST00000372978.3
ENST00000494547.1 ENST00000456846.2 ENST00000372982.4 ENST00000472818.1 ENST00000372977.3 |
TAF8
|
TAF8 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 43kDa |
| chr13_-_30169807 | 0.37 |
ENST00000380752.5
|
SLC7A1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr2_+_26915584 | 0.36 |
ENST00000302909.3
|
KCNK3
|
potassium channel, subfamily K, member 3 |
| chr16_-_755726 | 0.36 |
ENST00000324361.5
|
FBXL16
|
F-box and leucine-rich repeat protein 16 |
| chr20_+_44441304 | 0.36 |
ENST00000352551.5
|
UBE2C
|
ubiquitin-conjugating enzyme E2C |
| chr19_-_49121054 | 0.34 |
ENST00000546623.1
ENST00000084795.5 |
RPL18
|
ribosomal protein L18 |
| chr11_-_8985927 | 0.34 |
ENST00000528117.1
ENST00000309134.5 |
TMEM9B
|
TMEM9 domain family, member B |
| chr21_+_33784957 | 0.34 |
ENST00000401402.3
ENST00000382699.3 |
EVA1C
|
eva-1 homolog C (C. elegans) |
| chr6_-_36953833 | 0.33 |
ENST00000538808.1
ENST00000460219.1 ENST00000373616.5 ENST00000373627.5 |
MTCH1
|
mitochondrial carrier 1 |
| chr20_-_45061695 | 0.32 |
ENST00000445496.2
|
ELMO2
|
engulfment and cell motility 2 |
| chr21_-_45660723 | 0.32 |
ENST00000344330.4
ENST00000407780.3 ENST00000400379.3 |
ICOSLG
|
inducible T-cell co-stimulator ligand |
| chr5_-_137071756 | 0.31 |
ENST00000394937.3
ENST00000309755.4 |
KLHL3
|
kelch-like family member 3 |
| chr11_+_34460447 | 0.31 |
ENST00000241052.4
|
CAT
|
catalase |
Network of associatons between targets according to the STRING database.
First level regulatory network of GCM1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 4.8 | GO:0038043 | interleukin-5-mediated signaling pathway(GO:0038043) interleukin-3-mediated signaling pathway(GO:0038156) |
| 0.9 | 2.8 | GO:1900075 | regulation of neuromuscular synaptic transmission(GO:1900073) positive regulation of neuromuscular synaptic transmission(GO:1900075) |
| 0.8 | 2.4 | GO:0046168 | glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.6 | 0.6 | GO:1902304 | positive regulation of potassium ion export(GO:1902304) |
| 0.6 | 2.3 | GO:0002432 | granuloma formation(GO:0002432) |
| 0.5 | 1.5 | GO:0002522 | leukocyte migration involved in immune response(GO:0002522) |
| 0.3 | 0.9 | GO:0060139 | positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.3 | 0.9 | GO:0038162 | erythropoietin-mediated signaling pathway(GO:0038162) |
| 0.3 | 0.9 | GO:1904784 | NLRP1 inflammasome complex assembly(GO:1904784) |
| 0.3 | 2.6 | GO:0039663 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.3 | 0.9 | GO:0001812 | positive regulation of type IIa hypersensitivity(GO:0001798) positive regulation of type I hypersensitivity(GO:0001812) positive regulation of type II hypersensitivity(GO:0002894) positive regulation of mast cell cytokine production(GO:0032765) mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) |
| 0.3 | 0.9 | GO:2000308 | iron assimilation(GO:0033212) iron assimilation by chelation and transport(GO:0033214) positive regulation of bone mineralization involved in bone maturation(GO:1900159) negative regulation of tumor necrosis factor (ligand) superfamily member 11 production(GO:2000308) |
| 0.3 | 1.4 | GO:0008611 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.3 | 1.6 | GO:0002803 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.3 | 1.5 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.2 | 1.2 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.2 | 1.3 | GO:0045348 | positive regulation of MHC class II biosynthetic process(GO:0045348) |
| 0.2 | 30.8 | GO:0030449 | regulation of complement activation(GO:0030449) |
| 0.2 | 0.9 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.2 | 0.5 | GO:0007497 | posterior midgut development(GO:0007497) endothelin receptor signaling pathway(GO:0086100) |
| 0.2 | 0.7 | GO:0036071 | N-glycan fucosylation(GO:0036071) |
| 0.2 | 0.8 | GO:0038155 | interleukin-23-mediated signaling pathway(GO:0038155) |
| 0.2 | 0.8 | GO:0097021 | lymphocyte migration into lymphoid organs(GO:0097021) |
| 0.1 | 6.4 | GO:0035590 | purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.1 | 32.1 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.1 | 4.2 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.1 | 2.2 | GO:0043306 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.1 | 1.0 | GO:1900623 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.1 | 0.8 | GO:0002826 | negative regulation of T-helper 1 type immune response(GO:0002826) interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.1 | 3.5 | GO:0045589 | regulation of regulatory T cell differentiation(GO:0045589) |
| 0.1 | 0.7 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.1 | 0.7 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.1 | 0.4 | GO:0003335 | corneocyte development(GO:0003335) |
| 0.1 | 0.6 | GO:0007602 | phototransduction(GO:0007602) |
| 0.1 | 0.5 | GO:1902724 | mineralocorticoid receptor signaling pathway(GO:0031959) positive regulation of skeletal muscle satellite cell proliferation(GO:1902724) positive regulation of growth factor dependent skeletal muscle satellite cell proliferation(GO:1902728) |
| 0.1 | 0.3 | GO:0033320 | UDP-D-xylose metabolic process(GO:0033319) UDP-D-xylose biosynthetic process(GO:0033320) |
| 0.1 | 2.2 | GO:2000188 | regulation of cholesterol homeostasis(GO:2000188) |
| 0.1 | 2.4 | GO:0034162 | toll-like receptor 9 signaling pathway(GO:0034162) |
| 0.1 | 0.8 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.1 | 0.6 | GO:0098989 | NMDA selective glutamate receptor signaling pathway(GO:0098989) |
| 0.1 | 0.9 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 0.1 | 0.7 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.1 | 1.4 | GO:0002710 | negative regulation of T cell mediated immunity(GO:0002710) |
| 0.1 | 3.0 | GO:0043029 | T cell homeostasis(GO:0043029) |
| 0.1 | 0.4 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.1 | 1.1 | GO:0038166 | angiotensin-activated signaling pathway(GO:0038166) |
| 0.1 | 0.2 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) |
| 0.1 | 0.3 | GO:0010193 | response to ozone(GO:0010193) response to L-ascorbic acid(GO:0033591) |
| 0.0 | 1.3 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 1.8 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.3 | GO:0072156 | distal tubule morphogenesis(GO:0072156) |
| 0.0 | 0.2 | GO:0072086 | specification of loop of Henle identity(GO:0072086) pattern specification involved in metanephros development(GO:0072268) |
| 0.0 | 0.3 | GO:0071603 | endothelial cell-cell adhesion(GO:0071603) |
| 0.0 | 0.2 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.0 | 0.4 | GO:1903826 | arginine transmembrane transport(GO:1903826) |
| 0.0 | 1.0 | GO:0031065 | positive regulation of histone deacetylation(GO:0031065) |
| 0.0 | 0.3 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.2 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.0 | 0.5 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.0 | 0.4 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.7 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.0 | 0.6 | GO:0014894 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 0.4 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 0.6 | GO:2001204 | regulation of osteoclast development(GO:2001204) |
| 0.0 | 0.8 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.2 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.0 | 2.1 | GO:0002292 | T cell differentiation involved in immune response(GO:0002292) |
| 0.0 | 0.2 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.0 | 0.1 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 0.0 | 0.3 | GO:0046185 | aldehyde catabolic process(GO:0046185) |
| 0.0 | 0.6 | GO:0090022 | regulation of neutrophil chemotaxis(GO:0090022) |
| 0.0 | 2.1 | GO:0071349 | interleukin-12-mediated signaling pathway(GO:0035722) cellular response to interleukin-12(GO:0071349) |
| 0.0 | 0.2 | GO:1902363 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 0.0 | 0.5 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.2 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.4 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.2 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 1.4 | GO:0070266 | necroptotic process(GO:0070266) |
| 0.0 | 0.9 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 1.2 | GO:0008333 | endosome to lysosome transport(GO:0008333) |
| 0.0 | 0.2 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.0 | 1.0 | GO:0045576 | mast cell activation(GO:0045576) |
| 0.0 | 0.2 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 | 0.4 | GO:0051481 | negative regulation of cytosolic calcium ion concentration(GO:0051481) |
| 0.0 | 0.7 | GO:0032731 | positive regulation of interleukin-1 beta production(GO:0032731) |
| 0.0 | 0.4 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.0 | 0.3 | GO:0042104 | positive regulation of activated T cell proliferation(GO:0042104) |
| 0.0 | 0.1 | GO:0097084 | vascular smooth muscle cell development(GO:0097084) |
| 0.0 | 0.6 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 1.5 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 5.2 | GO:0002250 | adaptive immune response(GO:0002250) |
| 0.0 | 0.1 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.0 | 0.5 | GO:0051436 | negative regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle(GO:0051436) |
| 0.0 | 0.9 | GO:0046579 | positive regulation of Ras protein signal transduction(GO:0046579) |
| 0.0 | 0.2 | GO:0048147 | negative regulation of fibroblast proliferation(GO:0048147) |
| 0.0 | 0.1 | GO:0042182 | ketone catabolic process(GO:0042182) |
| 0.0 | 2.1 | GO:0051056 | regulation of small GTPase mediated signal transduction(GO:0051056) |
| 0.0 | 0.3 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 0.2 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.0 | 0.3 | GO:0045161 | neuronal ion channel clustering(GO:0045161) |
| 0.0 | 0.5 | GO:0006379 | mRNA cleavage(GO:0006379) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.8 | GO:0030526 | granulocyte macrophage colony-stimulating factor receptor complex(GO:0030526) |
| 0.4 | 1.2 | GO:0008623 | CHRAC(GO:0008623) |
| 0.3 | 2.4 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.3 | 0.9 | GO:0032998 | Fc receptor complex(GO:0032997) Fc-epsilon receptor I complex(GO:0032998) |
| 0.2 | 6.4 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.2 | 1.0 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.2 | 2.8 | GO:0099634 | postsynaptic specialization membrane(GO:0099634) |
| 0.2 | 0.8 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 0.1 | 1.6 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.1 | 0.9 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.1 | 1.0 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.1 | 0.9 | GO:0097013 | phagocytic vesicle lumen(GO:0097013) |
| 0.1 | 2.3 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.1 | 0.4 | GO:0097196 | Shu complex(GO:0097196) |
| 0.1 | 0.8 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.1 | 0.3 | GO:0071665 | gamma-catenin-TCF7L2 complex(GO:0071665) |
| 0.1 | 1.3 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 0.9 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.1 | 0.2 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.8 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 2.5 | GO:0031091 | platelet alpha granule(GO:0031091) |
| 0.0 | 0.4 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 1.2 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 8.0 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.6 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 2.2 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.6 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.7 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 1.5 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.5 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.2 | GO:0055028 | cortical microtubule(GO:0055028) |
| 0.0 | 0.8 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.1 | GO:1903440 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.0 | 2.1 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.0 | 0.5 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 1.5 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 41.7 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.2 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.9 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.0 | 3.1 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 1.9 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 0.3 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 1.0 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.2 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 1.6 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 1.9 | GO:0016605 | PML body(GO:0016605) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 4.8 | GO:0004912 | interleukin-3 receptor activity(GO:0004912) interleukin-5 receptor activity(GO:0004914) |
| 0.8 | 2.4 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.5 | 6.4 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.4 | 2.1 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.3 | 2.2 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.3 | 0.8 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.2 | 1.2 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.2 | 1.4 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.2 | 58.2 | GO:0003823 | antigen binding(GO:0003823) |
| 0.2 | 0.6 | GO:0050135 | NAD(P)+ nucleosidase activity(GO:0050135) |
| 0.2 | 1.5 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.2 | 2.6 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.2 | 0.9 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.2 | 0.5 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.2 | 0.7 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.2 | 0.7 | GO:0008424 | glycoprotein 6-alpha-L-fucosyltransferase activity(GO:0008424) alpha-(1->6)-fucosyltransferase activity(GO:0046921) |
| 0.2 | 0.8 | GO:0042019 | interleukin-23 binding(GO:0042019) interleukin-23 receptor activity(GO:0042020) |
| 0.2 | 1.1 | GO:0001595 | angiotensin receptor activity(GO:0001595) angiotensin type II receptor activity(GO:0004945) |
| 0.1 | 0.9 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.1 | 3.4 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.1 | 0.6 | GO:0047685 | amine sulfotransferase activity(GO:0047685) |
| 0.1 | 3.3 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.1 | 0.3 | GO:0048040 | UDP-glucuronate decarboxylase activity(GO:0048040) |
| 0.1 | 1.1 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.1 | 0.3 | GO:0005199 | structural constituent of cell wall(GO:0005199) |
| 0.1 | 0.8 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.1 | 0.2 | GO:0038131 | neuregulin receptor activity(GO:0038131) |
| 0.1 | 0.7 | GO:0004645 | phosphorylase activity(GO:0004645) |
| 0.1 | 0.3 | GO:0008267 | poly-glutamine tract binding(GO:0008267) |
| 0.1 | 0.3 | GO:0004096 | catalase activity(GO:0004096) |
| 0.1 | 0.2 | GO:0008441 | 3'(2'),5'-bisphosphate nucleotidase activity(GO:0008441) |
| 0.0 | 0.4 | GO:0004883 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.0 | 1.1 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 1.6 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.0 | 0.8 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 1.8 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.0 | 1.3 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 1.0 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.9 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.0 | 1.0 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 2.4 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 1.2 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.2 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.1 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 0.2 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.0 | 0.2 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.7 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.5 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 1.5 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.3 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.0 | 1.2 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.3 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 3.5 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 0.1 | GO:0097643 | amylin receptor activity(GO:0097643) |
| 0.0 | 0.5 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.6 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.2 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.6 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 0.3 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 5.6 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 0.4 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.6 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.5 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 1.2 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.3 | GO:0005351 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) |
| 0.0 | 0.2 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.0 | 0.6 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.2 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 1.6 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 1.3 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.3 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.1 | 6.3 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.1 | 0.8 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.1 | 1.6 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 2.4 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 3.3 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 1.0 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 6.3 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 1.2 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 1.0 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.8 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 1.5 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.7 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.9 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 0.7 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 1.2 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.9 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.4 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 3.2 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.3 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.6 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.6 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.2 | 2.4 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.1 | 5.3 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.1 | 3.9 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 2.2 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.1 | 0.9 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 1.5 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.1 | 2.3 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 1.4 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.8 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 1.0 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 1.4 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.9 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.9 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 1.1 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.4 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.0 | 0.6 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 1.0 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 0.6 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.5 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.5 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.9 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 1.1 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 1.2 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.7 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 1.3 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.3 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.4 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.8 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 1.3 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 1.0 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 0.3 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.4 | REACTOME AUTODEGRADATION OF CDH1 BY CDH1 APC C | Genes involved in Autodegradation of Cdh1 by Cdh1:APC/C |