Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
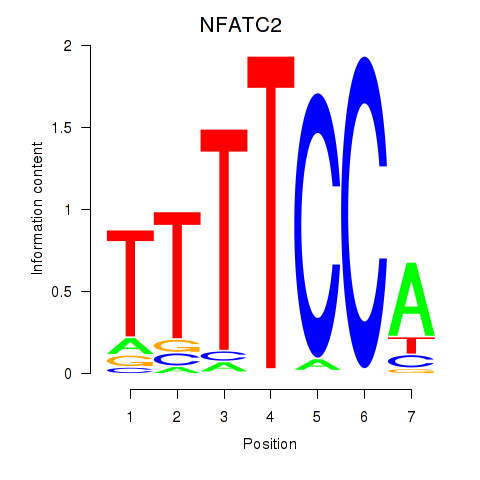
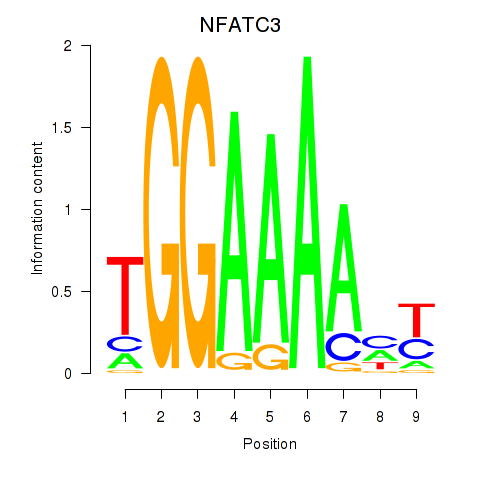
Results for NFATC2_NFATC3
Z-value: 1.50
Transcription factors associated with NFATC2_NFATC3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NFATC2
|
ENSG00000101096.15 | nuclear factor of activated T cells 2 |
|
NFATC3
|
ENSG00000072736.14 | nuclear factor of activated T cells 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NFATC3 | hg19_v2_chr16_+_68119247_68119293 | -0.67 | 8.4e-30 | Click! |
Activity profile of NFATC2_NFATC3 motif
Sorted Z-values of NFATC2_NFATC3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_91573132 | 44.23 |
ENST00000550563.1
ENST00000546370.1 |
DCN
|
decorin |
| chr12_-_91573249 | 41.61 |
ENST00000550099.1
ENST00000546391.1 ENST00000551354.1 |
DCN
|
decorin |
| chr12_-_91573316 | 34.23 |
ENST00000393155.1
|
DCN
|
decorin |
| chr2_-_238322770 | 29.29 |
ENST00000472056.1
|
COL6A3
|
collagen, type VI, alpha 3 |
| chr9_-_35691017 | 28.54 |
ENST00000378292.3
|
TPM2
|
tropomyosin 2 (beta) |
| chr1_+_86046433 | 28.07 |
ENST00000451137.2
|
CYR61
|
cysteine-rich, angiogenic inducer, 61 |
| chr2_-_238322800 | 26.80 |
ENST00000392004.3
ENST00000433762.1 ENST00000347401.3 ENST00000353578.4 ENST00000346358.4 ENST00000392003.2 |
COL6A3
|
collagen, type VI, alpha 3 |
| chr12_-_91539918 | 26.62 |
ENST00000548218.1
|
DCN
|
decorin |
| chr2_-_238323007 | 24.07 |
ENST00000295550.4
|
COL6A3
|
collagen, type VI, alpha 3 |
| chr14_-_92413353 | 23.58 |
ENST00000556154.1
|
FBLN5
|
fibulin 5 |
| chr12_-_9268707 | 23.28 |
ENST00000318602.7
|
A2M
|
alpha-2-macroglobulin |
| chr14_-_92413727 | 20.69 |
ENST00000267620.10
|
FBLN5
|
fibulin 5 |
| chr17_+_20059302 | 19.65 |
ENST00000395530.2
|
SPECC1
|
sperm antigen with calponin homology and coiled-coil domains 1 |
| chr12_-_50616122 | 19.37 |
ENST00000552823.1
ENST00000552909.1 |
LIMA1
|
LIM domain and actin binding 1 |
| chr8_-_13134045 | 16.66 |
ENST00000512044.2
|
DLC1
|
deleted in liver cancer 1 |
| chr12_-_50616382 | 15.96 |
ENST00000552783.1
|
LIMA1
|
LIM domain and actin binding 1 |
| chr2_-_163099885 | 14.98 |
ENST00000443424.1
|
FAP
|
fibroblast activation protein, alpha |
| chr2_-_163100045 | 14.65 |
ENST00000188790.4
|
FAP
|
fibroblast activation protein, alpha |
| chr2_-_190044480 | 14.62 |
ENST00000374866.3
|
COL5A2
|
collagen, type V, alpha 2 |
| chr16_-_3306587 | 14.58 |
ENST00000541159.1
ENST00000536379.1 ENST00000219596.1 ENST00000339854.4 |
MEFV
|
Mediterranean fever |
| chr7_+_20686946 | 14.21 |
ENST00000443026.2
ENST00000406935.1 |
ABCB5
|
ATP-binding cassette, sub-family B (MDR/TAP), member 5 |
| chr3_-_114035026 | 13.85 |
ENST00000570269.1
|
RP11-553L6.5
|
RP11-553L6.5 |
| chr9_+_124062071 | 13.57 |
ENST00000373818.4
|
GSN
|
gelsolin |
| chr11_-_111794446 | 13.50 |
ENST00000527950.1
|
CRYAB
|
crystallin, alpha B |
| chr19_+_41509851 | 13.08 |
ENST00000593831.1
ENST00000330446.5 |
CYP2B6
|
cytochrome P450, family 2, subfamily B, polypeptide 6 |
| chr3_-_195310802 | 12.79 |
ENST00000421243.1
ENST00000453131.1 |
APOD
|
apolipoprotein D |
| chr15_-_90892669 | 12.53 |
ENST00000412799.2
|
GABARAPL3
|
GABA(A) receptors associated protein like 3, pseudogene |
| chr5_-_42811986 | 12.32 |
ENST00000511224.1
ENST00000507920.1 ENST00000510965.1 |
SEPP1
|
selenoprotein P, plasma, 1 |
| chr8_+_98900132 | 11.88 |
ENST00000520016.1
|
MATN2
|
matrilin 2 |
| chr5_-_42812143 | 11.83 |
ENST00000514985.1
|
SEPP1
|
selenoprotein P, plasma, 1 |
| chr15_+_93443419 | 11.75 |
ENST00000557381.1
ENST00000420239.2 |
CHD2
|
chromodomain helicase DNA binding protein 2 |
| chr8_-_95274536 | 11.67 |
ENST00000297596.2
ENST00000396194.2 |
GEM
|
GTP binding protein overexpressed in skeletal muscle |
| chr5_+_135394840 | 11.47 |
ENST00000503087.1
|
TGFBI
|
transforming growth factor, beta-induced, 68kDa |
| chr7_+_134551583 | 10.95 |
ENST00000435928.1
|
CALD1
|
caldesmon 1 |
| chr11_-_119293872 | 10.91 |
ENST00000524970.1
|
THY1
|
Thy-1 cell surface antigen |
| chr3_-_183273477 | 10.57 |
ENST00000341319.3
|
KLHL6
|
kelch-like family member 6 |
| chr19_+_37998031 | 10.36 |
ENST00000586138.1
ENST00000588578.1 ENST00000587986.1 |
ZNF793
|
zinc finger protein 793 |
| chr13_-_88323218 | 10.03 |
ENST00000436290.2
ENST00000453832.2 ENST00000606590.1 |
MIR4500HG
|
MIR4500 host gene (non-protein coding) |
| chr16_+_15528332 | 9.99 |
ENST00000566490.1
|
C16orf45
|
chromosome 16 open reading frame 45 |
| chr21_+_27011584 | 9.82 |
ENST00000400532.1
ENST00000480456.1 ENST00000312957.5 |
JAM2
|
junctional adhesion molecule 2 |
| chr15_+_63340647 | 9.74 |
ENST00000404484.4
|
TPM1
|
tropomyosin 1 (alpha) |
| chr3_-_123512688 | 9.52 |
ENST00000475616.1
|
MYLK
|
myosin light chain kinase |
| chr17_-_1389228 | 9.24 |
ENST00000438665.2
|
MYO1C
|
myosin IC |
| chr15_+_63340858 | 9.02 |
ENST00000560615.1
|
TPM1
|
tropomyosin 1 (alpha) |
| chr22_-_38380543 | 8.94 |
ENST00000396884.2
|
SOX10
|
SRY (sex determining region Y)-box 10 |
| chr17_-_8263538 | 8.92 |
ENST00000535173.1
|
AC135178.1
|
HCG1985372; Uncharacterized protein; cDNA FLJ37541 fis, clone BRCAN2026340 |
| chr8_+_80523321 | 8.85 |
ENST00000518111.1
|
STMN2
|
stathmin-like 2 |
| chr15_+_96869165 | 8.82 |
ENST00000421109.2
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr15_-_65503801 | 8.70 |
ENST00000261883.4
|
CILP
|
cartilage intermediate layer protein, nucleotide pyrophosphohydrolase |
| chr1_-_150780757 | 8.65 |
ENST00000271651.3
|
CTSK
|
cathepsin K |
| chr15_+_63334831 | 8.63 |
ENST00000288398.6
ENST00000358278.3 ENST00000560445.1 ENST00000403994.3 ENST00000357980.4 ENST00000559556.1 ENST00000559397.1 ENST00000267996.7 ENST00000560970.1 |
TPM1
|
tropomyosin 1 (alpha) |
| chr6_+_31795506 | 8.59 |
ENST00000375650.3
|
HSPA1B
|
heat shock 70kDa protein 1B |
| chr12_-_95467267 | 8.54 |
ENST00000330677.7
|
NR2C1
|
nuclear receptor subfamily 2, group C, member 1 |
| chr5_+_140186647 | 8.47 |
ENST00000512229.2
ENST00000356878.4 ENST00000530339.1 |
PCDHA4
|
protocadherin alpha 4 |
| chrX_+_22056165 | 8.43 |
ENST00000535894.1
|
PHEX
|
phosphate regulating endopeptidase homolog, X-linked |
| chr19_+_58095501 | 8.35 |
ENST00000536878.2
ENST00000597850.1 ENST00000597219.1 ENST00000598689.1 ENST00000599456.1 ENST00000307468.4 |
ZIK1
|
zinc finger protein interacting with K protein 1 |
| chr15_-_37392086 | 8.33 |
ENST00000561208.1
|
MEIS2
|
Meis homeobox 2 |
| chr15_+_63340734 | 8.28 |
ENST00000560959.1
|
TPM1
|
tropomyosin 1 (alpha) |
| chr6_+_151662815 | 8.14 |
ENST00000359755.5
|
AKAP12
|
A kinase (PRKA) anchor protein 12 |
| chr8_-_110703819 | 8.14 |
ENST00000532779.1
ENST00000534578.1 |
SYBU
|
syntabulin (syntaxin-interacting) |
| chr1_+_162602244 | 7.97 |
ENST00000367922.3
ENST00000367921.3 |
DDR2
|
discoidin domain receptor tyrosine kinase 2 |
| chr17_-_1389419 | 7.96 |
ENST00000575158.1
|
MYO1C
|
myosin IC |
| chrX_+_56259316 | 7.90 |
ENST00000468660.1
|
KLF8
|
Kruppel-like factor 8 |
| chr10_+_128593978 | 7.87 |
ENST00000280333.6
|
DOCK1
|
dedicator of cytokinesis 1 |
| chr17_+_59477233 | 7.79 |
ENST00000240328.3
|
TBX2
|
T-box 2 |
| chr8_-_120685608 | 7.57 |
ENST00000427067.2
|
ENPP2
|
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
| chr16_+_31483451 | 7.54 |
ENST00000565360.1
ENST00000361773.3 |
TGFB1I1
|
transforming growth factor beta 1 induced transcript 1 |
| chr8_+_70404996 | 7.50 |
ENST00000402687.4
ENST00000419716.3 |
SULF1
|
sulfatase 1 |
| chr1_+_244816237 | 7.38 |
ENST00000302550.11
|
DESI2
|
desumoylating isopeptidase 2 |
| chrX_-_17878827 | 7.28 |
ENST00000360011.1
|
RAI2
|
retinoic acid induced 2 |
| chr4_+_41614909 | 7.14 |
ENST00000509454.1
ENST00000396595.3 ENST00000381753.4 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr19_-_36523709 | 7.13 |
ENST00000592017.1
ENST00000360535.4 |
CLIP3
|
CAP-GLY domain containing linker protein 3 |
| chrY_-_15591485 | 7.07 |
ENST00000382896.4
ENST00000537580.1 ENST00000540140.1 ENST00000545955.1 ENST00000538878.1 |
UTY
|
ubiquitously transcribed tetratricopeptide repeat containing, Y-linked |
| chr19_+_11658655 | 7.06 |
ENST00000588935.1
|
CNN1
|
calponin 1, basic, smooth muscle |
| chr17_-_38256973 | 6.97 |
ENST00000246672.3
|
NR1D1
|
nuclear receptor subfamily 1, group D, member 1 |
| chr14_+_24837226 | 6.88 |
ENST00000554050.1
ENST00000554903.1 ENST00000554779.1 ENST00000250373.4 ENST00000553708.1 |
NFATC4
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4 |
| chr14_-_52535712 | 6.84 |
ENST00000216286.5
ENST00000541773.1 |
NID2
|
nidogen 2 (osteonidogen) |
| chr12_+_12938541 | 6.74 |
ENST00000356591.4
|
APOLD1
|
apolipoprotein L domain containing 1 |
| chr4_+_55095264 | 6.72 |
ENST00000257290.5
|
PDGFRA
|
platelet-derived growth factor receptor, alpha polypeptide |
| chr18_+_23806437 | 6.71 |
ENST00000578121.1
|
TAF4B
|
TAF4b RNA polymerase II, TATA box binding protein (TBP)-associated factor, 105kDa |
| chr5_+_140220769 | 6.68 |
ENST00000531613.1
ENST00000378123.3 |
PCDHA8
|
protocadherin alpha 8 |
| chr16_+_55542910 | 6.65 |
ENST00000262134.5
|
LPCAT2
|
lysophosphatidylcholine acyltransferase 2 |
| chr3_+_148447887 | 6.63 |
ENST00000475347.1
ENST00000474935.1 ENST00000461609.1 |
AGTR1
|
angiotensin II receptor, type 1 |
| chr4_+_41258786 | 6.61 |
ENST00000503431.1
ENST00000284440.4 ENST00000508768.1 ENST00000512788.1 |
UCHL1
|
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase) |
| chr16_-_71610985 | 6.53 |
ENST00000355962.4
|
TAT
|
tyrosine aminotransferase |
| chr15_+_63354769 | 6.53 |
ENST00000558910.1
|
TPM1
|
tropomyosin 1 (alpha) |
| chr5_-_77844974 | 6.52 |
ENST00000515007.2
|
LHFPL2
|
lipoma HMGIC fusion partner-like 2 |
| chr4_-_168155300 | 6.47 |
ENST00000541637.1
|
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr8_-_110704014 | 6.45 |
ENST00000529190.1
ENST00000422135.1 ENST00000419099.1 |
SYBU
|
syntabulin (syntaxin-interacting) |
| chr15_+_63340775 | 6.45 |
ENST00000559281.1
ENST00000317516.7 |
TPM1
|
tropomyosin 1 (alpha) |
| chr11_+_75273246 | 6.44 |
ENST00000526397.1
ENST00000529643.1 ENST00000525492.1 ENST00000530284.1 ENST00000532356.1 ENST00000524558.1 |
SERPINH1
|
serpin peptidase inhibitor, clade H (heat shock protein 47), member 1, (collagen binding protein 1) |
| chr12_+_10365404 | 6.44 |
ENST00000266458.5
ENST00000421801.2 ENST00000544284.1 ENST00000545047.1 ENST00000543602.1 ENST00000545887.1 |
GABARAPL1
|
GABA(A) receptor-associated protein like 1 |
| chr6_-_52859046 | 6.44 |
ENST00000457564.1
ENST00000541324.1 ENST00000370960.1 |
GSTA4
|
glutathione S-transferase alpha 4 |
| chr5_+_140772381 | 6.41 |
ENST00000398604.2
|
PCDHGA8
|
protocadherin gamma subfamily A, 8 |
| chr22_-_19512893 | 6.36 |
ENST00000403084.1
ENST00000413119.2 |
CLDN5
|
claudin 5 |
| chr9_+_103204553 | 6.29 |
ENST00000502978.1
ENST00000334943.6 |
MSANTD3-TMEFF1
TMEFF1
|
MSANTD3-TMEFF1 readthrough transmembrane protein with EGF-like and two follistatin-like domains 1 |
| chr6_+_86159821 | 6.27 |
ENST00000369651.3
|
NT5E
|
5'-nucleotidase, ecto (CD73) |
| chr1_+_46640750 | 6.22 |
ENST00000372003.1
|
TSPAN1
|
tetraspanin 1 |
| chr2_+_46926326 | 6.01 |
ENST00000394861.2
|
SOCS5
|
suppressor of cytokine signaling 5 |
| chr5_+_131993856 | 5.91 |
ENST00000304506.3
|
IL13
|
interleukin 13 |
| chr11_+_75273101 | 5.90 |
ENST00000533603.1
ENST00000358171.3 ENST00000526242.1 |
SERPINH1
|
serpin peptidase inhibitor, clade H (heat shock protein 47), member 1, (collagen binding protein 1) |
| chr16_+_56642041 | 5.86 |
ENST00000245185.5
|
MT2A
|
metallothionein 2A |
| chr6_-_11779840 | 5.82 |
ENST00000506810.1
|
ADTRP
|
androgen-dependent TFPI-regulating protein |
| chr10_+_74451883 | 5.81 |
ENST00000373053.3
ENST00000357157.6 |
MCU
|
mitochondrial calcium uniporter |
| chr2_+_234580525 | 5.73 |
ENST00000609637.1
|
UGT1A1
|
UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr14_+_32546274 | 5.70 |
ENST00000396582.2
|
ARHGAP5
|
Rho GTPase activating protein 5 |
| chr7_+_150497491 | 5.67 |
ENST00000484928.1
|
TMEM176A
|
transmembrane protein 176A |
| chr14_+_65171099 | 5.61 |
ENST00000247226.7
|
PLEKHG3
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
| chr12_-_104443890 | 5.57 |
ENST00000547583.1
ENST00000360814.4 ENST00000546851.1 |
GLT8D2
|
glycosyltransferase 8 domain containing 2 |
| chr5_+_140864649 | 5.57 |
ENST00000306593.1
|
PCDHGC4
|
protocadherin gamma subfamily C, 4 |
| chr6_+_43739697 | 5.56 |
ENST00000230480.6
|
VEGFA
|
vascular endothelial growth factor A |
| chr22_-_36220420 | 5.55 |
ENST00000473487.2
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr6_+_86159765 | 5.54 |
ENST00000369646.3
ENST00000257770.3 |
NT5E
|
5'-nucleotidase, ecto (CD73) |
| chr1_+_179712298 | 5.49 |
ENST00000341785.4
|
FAM163A
|
family with sequence similarity 163, member A |
| chr17_+_74372662 | 5.47 |
ENST00000591651.1
ENST00000545180.1 |
SPHK1
|
sphingosine kinase 1 |
| chr20_+_19867150 | 5.46 |
ENST00000255006.6
|
RIN2
|
Ras and Rab interactor 2 |
| chr16_+_66400533 | 5.44 |
ENST00000341529.3
|
CDH5
|
cadherin 5, type 2 (vascular endothelium) |
| chr16_-_66952742 | 5.32 |
ENST00000565235.2
ENST00000568632.1 ENST00000565796.1 |
CDH16
|
cadherin 16, KSP-cadherin |
| chr1_-_151319283 | 5.25 |
ENST00000392746.3
|
RFX5
|
regulatory factor X, 5 (influences HLA class II expression) |
| chr14_+_75536335 | 5.22 |
ENST00000554763.1
ENST00000439583.2 ENST00000526130.1 ENST00000525046.1 |
ZC2HC1C
|
zinc finger, C2HC-type containing 1C |
| chr9_+_35538616 | 5.18 |
ENST00000455600.1
|
RUSC2
|
RUN and SH3 domain containing 2 |
| chr1_+_54519242 | 5.18 |
ENST00000234827.1
|
TCEANC2
|
transcription elongation factor A (SII) N-terminal and central domain containing 2 |
| chrX_-_37706661 | 5.15 |
ENST00000432389.2
ENST00000378581.3 |
DYNLT3
|
dynein, light chain, Tctex-type 3 |
| chr4_+_169418195 | 5.14 |
ENST00000261509.6
ENST00000335742.7 |
PALLD
|
palladin, cytoskeletal associated protein |
| chrX_+_84498989 | 5.14 |
ENST00000395402.1
|
ZNF711
|
zinc finger protein 711 |
| chr14_+_75536280 | 5.12 |
ENST00000238686.8
|
ZC2HC1C
|
zinc finger, C2HC-type containing 1C |
| chrX_-_40506766 | 4.97 |
ENST00000378421.1
ENST00000440784.2 ENST00000327877.5 ENST00000378426.1 ENST00000378418.2 |
CXorf38
|
chromosome X open reading frame 38 |
| chr5_+_132009675 | 4.96 |
ENST00000231449.2
ENST00000350025.2 |
IL4
|
interleukin 4 |
| chr9_+_19049372 | 4.94 |
ENST00000380527.1
|
RRAGA
|
Ras-related GTP binding A |
| chr5_-_138210977 | 4.91 |
ENST00000274711.6
ENST00000521094.2 |
LRRTM2
|
leucine rich repeat transmembrane neuronal 2 |
| chr20_+_36149602 | 4.86 |
ENST00000062104.2
ENST00000346199.2 |
NNAT
|
neuronatin |
| chr5_-_41261540 | 4.86 |
ENST00000263413.3
|
C6
|
complement component 6 |
| chr3_+_148415571 | 4.85 |
ENST00000497524.1
ENST00000349243.3 ENST00000542281.1 ENST00000418473.2 ENST00000404754.2 |
AGTR1
|
angiotensin II receptor, type 1 |
| chr12_-_16758059 | 4.82 |
ENST00000261169.6
|
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr3_+_142442841 | 4.80 |
ENST00000476941.1
ENST00000273482.6 |
TRPC1
|
transient receptor potential cation channel, subfamily C, member 1 |
| chr5_+_140762268 | 4.77 |
ENST00000518325.1
|
PCDHGA7
|
protocadherin gamma subfamily A, 7 |
| chr8_-_38853990 | 4.77 |
ENST00000456845.2
ENST00000397070.2 ENST00000517872.1 ENST00000412303.1 ENST00000456397.2 |
TM2D2
|
TM2 domain containing 2 |
| chr4_+_88571429 | 4.74 |
ENST00000339673.6
ENST00000282479.7 |
DMP1
|
dentin matrix acidic phosphoprotein 1 |
| chr16_+_29674540 | 4.72 |
ENST00000436527.1
ENST00000360121.3 ENST00000449759.1 |
SPN
QPRT
|
sialophorin quinolinate phosphoribosyltransferase |
| chr16_-_66952779 | 4.72 |
ENST00000570262.1
ENST00000394055.3 ENST00000299752.4 |
CDH16
|
cadherin 16, KSP-cadherin |
| chr1_-_68698222 | 4.70 |
ENST00000370976.3
ENST00000354777.2 ENST00000262348.4 ENST00000540432.1 |
WLS
|
wntless Wnt ligand secretion mediator |
| chr9_+_137533615 | 4.68 |
ENST00000371817.3
|
COL5A1
|
collagen, type V, alpha 1 |
| chrX_+_52780318 | 4.67 |
ENST00000375515.3
ENST00000276049.6 |
SSX2B
|
synovial sarcoma, X breakpoint 2B |
| chr12_-_16758304 | 4.66 |
ENST00000320122.6
|
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr18_-_500692 | 4.62 |
ENST00000400256.3
|
COLEC12
|
collectin sub-family member 12 |
| chr2_+_234580499 | 4.62 |
ENST00000354728.4
|
UGT1A9
|
UDP glucuronosyltransferase 1 family, polypeptide A9 |
| chr3_-_167813132 | 4.61 |
ENST00000309027.4
|
GOLIM4
|
golgi integral membrane protein 4 |
| chr10_+_105036909 | 4.55 |
ENST00000369849.4
|
INA
|
internexin neuronal intermediate filament protein, alpha |
| chr11_-_119187826 | 4.52 |
ENST00000264036.4
|
MCAM
|
melanoma cell adhesion molecule |
| chr14_-_89878369 | 4.52 |
ENST00000553840.1
ENST00000556916.1 |
FOXN3
|
forkhead box N3 |
| chr1_+_82266053 | 4.51 |
ENST00000370715.1
ENST00000370713.1 ENST00000319517.6 ENST00000370717.2 ENST00000394879.1 ENST00000271029.4 ENST00000335786.5 |
LPHN2
|
latrophilin 2 |
| chr2_+_11696464 | 4.50 |
ENST00000234142.5
|
GREB1
|
growth regulation by estrogen in breast cancer 1 |
| chr15_+_32933866 | 4.50 |
ENST00000300175.4
ENST00000413748.2 ENST00000494364.1 ENST00000497208.1 |
SCG5
|
secretogranin V (7B2 protein) |
| chr10_-_29923893 | 4.49 |
ENST00000355867.4
|
SVIL
|
supervillin |
| chr4_+_39408470 | 4.49 |
ENST00000257408.4
|
KLB
|
klotho beta |
| chr13_-_33780133 | 4.47 |
ENST00000399365.3
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13 |
| chr15_-_37391614 | 4.43 |
ENST00000219869.9
|
MEIS2
|
Meis homeobox 2 |
| chr17_-_34257731 | 4.43 |
ENST00000431884.2
ENST00000425909.3 ENST00000394528.3 ENST00000430160.2 |
RDM1
|
RAD52 motif 1 |
| chr15_+_74466744 | 4.39 |
ENST00000560862.1
ENST00000395118.1 |
ISLR
|
immunoglobulin superfamily containing leucine-rich repeat |
| chr4_-_186877806 | 4.38 |
ENST00000355634.5
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr2_-_206950781 | 4.38 |
ENST00000403263.1
|
INO80D
|
INO80 complex subunit D |
| chr2_-_227664474 | 4.35 |
ENST00000305123.5
|
IRS1
|
insulin receptor substrate 1 |
| chr1_-_94703118 | 4.34 |
ENST00000260526.6
ENST00000370217.3 |
ARHGAP29
|
Rho GTPase activating protein 29 |
| chr1_+_38022513 | 4.33 |
ENST00000296218.7
|
DNALI1
|
dynein, axonemal, light intermediate chain 1 |
| chr7_+_108210012 | 4.33 |
ENST00000249356.3
|
DNAJB9
|
DnaJ (Hsp40) homolog, subfamily B, member 9 |
| chr10_+_47746929 | 4.33 |
ENST00000340243.6
ENST00000374277.5 ENST00000449464.2 ENST00000538825.1 ENST00000335083.5 |
ANXA8L2
AL603965.1
|
annexin A8-like 2 Protein LOC100996760 |
| chr17_+_42925270 | 4.30 |
ENST00000253410.2
ENST00000587021.1 |
HIGD1B
|
HIG1 hypoxia inducible domain family, member 1B |
| chr7_-_92157760 | 4.30 |
ENST00000248633.4
|
PEX1
|
peroxisomal biogenesis factor 1 |
| chr11_-_76381029 | 4.29 |
ENST00000407242.2
ENST00000421973.1 |
LRRC32
|
leucine rich repeat containing 32 |
| chr1_-_109618566 | 4.25 |
ENST00000338366.5
|
TAF13
|
TAF13 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 18kDa |
| chr17_-_15522826 | 4.21 |
ENST00000395906.3
|
CDRT1
|
CMT1A duplicated region transcript 1 |
| chr6_-_134495992 | 4.20 |
ENST00000475719.2
ENST00000367857.5 ENST00000237305.7 |
SGK1
|
serum/glucocorticoid regulated kinase 1 |
| chr7_+_102553430 | 4.19 |
ENST00000339431.4
ENST00000249377.4 |
LRRC17
|
leucine rich repeat containing 17 |
| chr7_-_122526799 | 4.19 |
ENST00000334010.7
ENST00000313070.7 |
CADPS2
|
Ca++-dependent secretion activator 2 |
| chr15_+_43885252 | 4.17 |
ENST00000453782.1
ENST00000300283.6 ENST00000437924.1 ENST00000450086.2 |
CKMT1B
|
creatine kinase, mitochondrial 1B |
| chrX_+_37208540 | 4.15 |
ENST00000466533.1
ENST00000542554.1 ENST00000543642.1 ENST00000484460.1 ENST00000449135.2 ENST00000463135.1 ENST00000465127.1 |
PRRG1
TM4SF2
|
proline rich Gla (G-carboxyglutamic acid) 1 Uncharacterized protein; cDNA FLJ59144, highly similar to Tetraspanin-7 |
| chr11_+_64851729 | 4.14 |
ENST00000526791.1
ENST00000526945.1 |
ZFPL1
|
zinc finger protein-like 1 |
| chr17_-_39041479 | 4.12 |
ENST00000167588.3
|
KRT20
|
keratin 20 |
| chr5_+_140254884 | 4.09 |
ENST00000398631.2
|
PCDHA12
|
protocadherin alpha 12 |
| chr1_-_161207953 | 4.07 |
ENST00000367982.4
|
NR1I3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr6_+_31916733 | 4.05 |
ENST00000483004.1
|
CFB
|
complement factor B |
| chr12_+_54447637 | 4.04 |
ENST00000609810.1
ENST00000430889.2 |
HOXC4
HOXC4
|
homeobox C4 Homeobox protein Hox-C4 |
| chr6_-_131384373 | 4.03 |
ENST00000392427.3
ENST00000525271.1 ENST00000527411.1 |
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2 |
| chr18_-_52989217 | 4.02 |
ENST00000570287.2
|
TCF4
|
transcription factor 4 |
| chr9_-_13165457 | 4.01 |
ENST00000542239.1
ENST00000538841.1 ENST00000433359.2 |
MPDZ
|
multiple PDZ domain protein |
| chr6_-_62996066 | 4.01 |
ENST00000281156.4
|
KHDRBS2
|
KH domain containing, RNA binding, signal transduction associated 2 |
| chr22_-_36236265 | 4.00 |
ENST00000414461.2
ENST00000416721.2 ENST00000449924.2 ENST00000262829.7 ENST00000397305.3 |
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr22_-_36236623 | 3.99 |
ENST00000405409.2
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr1_+_215747118 | 3.97 |
ENST00000448333.1
|
KCTD3
|
potassium channel tetramerization domain containing 3 |
| chr7_+_120629653 | 3.95 |
ENST00000450913.2
ENST00000340646.5 |
CPED1
|
cadherin-like and PC-esterase domain containing 1 |
| chr11_-_10590238 | 3.95 |
ENST00000256178.3
|
LYVE1
|
lymphatic vessel endothelial hyaluronan receptor 1 |
| chr15_+_43985084 | 3.94 |
ENST00000434505.1
ENST00000411750.1 |
CKMT1A
|
creatine kinase, mitochondrial 1A |
| chr14_+_65171315 | 3.93 |
ENST00000394691.1
|
PLEKHG3
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
| chr11_+_22696314 | 3.92 |
ENST00000532398.1
ENST00000433790.1 |
GAS2
|
growth arrest-specific 2 |
| chr10_+_114710516 | 3.89 |
ENST00000542695.1
ENST00000346198.4 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chrX_+_44732757 | 3.87 |
ENST00000377967.4
ENST00000536777.1 ENST00000382899.4 ENST00000543216.1 |
KDM6A
|
lysine (K)-specific demethylase 6A |
| chr17_-_39093672 | 3.86 |
ENST00000209718.3
ENST00000436344.3 ENST00000485751.1 |
KRT23
|
keratin 23 (histone deacetylase inducible) |
| chr11_-_10590118 | 3.84 |
ENST00000529598.1
|
LYVE1
|
lymphatic vessel endothelial hyaluronan receptor 1 |
| chr12_-_76425368 | 3.84 |
ENST00000602540.1
|
PHLDA1
|
pleckstrin homology-like domain, family A, member 1 |
| chr7_+_127881325 | 3.84 |
ENST00000308868.4
|
LEP
|
leptin |
Network of associatons between targets according to the STRING database.
First level regulatory network of NFATC2_NFATC3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 12.2 | 146.7 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 7.4 | 29.6 | GO:0097325 | melanocyte proliferation(GO:0097325) |
| 6.1 | 48.6 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 4.9 | 14.6 | GO:1900226 | negative regulation of NLRP3 inflammasome complex assembly(GO:1900226) |
| 4.8 | 19.3 | GO:1903224 | regulation of endodermal cell differentiation(GO:1903224) |
| 4.3 | 12.8 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 3.9 | 11.7 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 3.7 | 44.3 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 3.6 | 10.9 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 3.6 | 10.9 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 3.6 | 14.2 | GO:0048749 | compound eye development(GO:0048749) |
| 3.1 | 28.1 | GO:0003278 | apoptotic process involved in heart morphogenesis(GO:0003278) |
| 3.0 | 11.8 | GO:0046086 | adenosine biosynthetic process(GO:0046086) |
| 2.9 | 11.5 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 2.8 | 11.3 | GO:2000320 | negative regulation of T-helper 17 type immune response(GO:2000317) negative regulation of T-helper 17 cell differentiation(GO:2000320) |
| 2.7 | 13.6 | GO:1903906 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
| 2.5 | 7.6 | GO:0033861 | negative regulation of NAD(P)H oxidase activity(GO:0033861) |
| 2.5 | 12.3 | GO:0038091 | VEGF-activated platelet-derived growth factor receptor signaling pathway(GO:0038086) positive regulation of cell proliferation by VEGF-activated platelet derived growth factor receptor signaling pathway(GO:0038091) |
| 2.4 | 14.7 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 2.4 | 7.3 | GO:0060151 | peroxisome localization(GO:0060151) microtubule-based peroxisome localization(GO:0060152) |
| 2.3 | 7.0 | GO:0060086 | circadian temperature homeostasis(GO:0060086) |
| 2.3 | 13.5 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 2.2 | 8.9 | GO:0071393 | cellular response to progesterone stimulus(GO:0071393) |
| 2.2 | 8.8 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 2.2 | 8.8 | GO:0009956 | radial pattern formation(GO:0009956) |
| 2.2 | 6.6 | GO:0007412 | axon target recognition(GO:0007412) |
| 2.1 | 8.6 | GO:0070434 | positive regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070426) positive regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070434) |
| 2.1 | 6.3 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 2.1 | 6.2 | GO:2000364 | regulation of STAT protein import into nucleus(GO:2000364) positive regulation of STAT protein import into nucleus(GO:2000366) |
| 1.9 | 7.5 | GO:0060686 | negative regulation of prostatic bud formation(GO:0060686) |
| 1.8 | 5.5 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 1.8 | 23.8 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 1.8 | 5.5 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 1.7 | 8.6 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 1.7 | 6.7 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 1.6 | 9.7 | GO:1990418 | response to insulin-like growth factor stimulus(GO:1990418) |
| 1.6 | 6.4 | GO:1904636 | response to ionomycin(GO:1904636) cellular response to ionomycin(GO:1904637) |
| 1.6 | 9.5 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 1.6 | 14.2 | GO:0051552 | flavone metabolic process(GO:0051552) |
| 1.6 | 7.8 | GO:0007521 | muscle cell fate determination(GO:0007521) |
| 1.6 | 6.2 | GO:0019249 | lactate biosynthetic process(GO:0019249) |
| 1.5 | 12.3 | GO:0003433 | chondrocyte development involved in endochondral bone morphogenesis(GO:0003433) |
| 1.5 | 7.6 | GO:1903976 | negative regulation of glial cell migration(GO:1903976) |
| 1.5 | 16.7 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 1.5 | 11.8 | GO:1901552 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 1.5 | 5.9 | GO:0010273 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 1.4 | 7.1 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 1.4 | 8.5 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 1.3 | 3.8 | GO:0045210 | FasL biosynthetic process(GO:0045210) |
| 1.3 | 3.8 | GO:0043006 | activation of phospholipase A2 activity by calcium-mediated signaling(GO:0043006) |
| 1.2 | 3.7 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 1.2 | 4.9 | GO:0001970 | positive regulation of activation of membrane attack complex(GO:0001970) |
| 1.2 | 9.5 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 1.2 | 4.7 | GO:0002884 | regulation of type IV hypersensitivity(GO:0001807) negative regulation of hypersensitivity(GO:0002884) |
| 1.2 | 87.8 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 1.1 | 2.3 | GO:0045168 | cell-cell signaling involved in cell fate commitment(GO:0045168) |
| 1.1 | 4.3 | GO:0021564 | vagus nerve development(GO:0021564) |
| 1.0 | 4.2 | GO:0048539 | bone marrow development(GO:0048539) |
| 1.0 | 3.1 | GO:0044278 | cell wall disruption in other organism(GO:0044278) |
| 1.0 | 12.4 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 1.0 | 4.1 | GO:0003409 | optic cup structural organization(GO:0003409) |
| 1.0 | 8.0 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 1.0 | 8.8 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 1.0 | 3.9 | GO:2000820 | negative regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000820) |
| 1.0 | 13.7 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 1.0 | 10.7 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 1.0 | 5.8 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 1.0 | 6.8 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 1.0 | 1.0 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) |
| 0.9 | 7.5 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.9 | 13.1 | GO:0042738 | exogenous drug catabolic process(GO:0042738) |
| 0.9 | 15.8 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.9 | 3.7 | GO:0009386 | translational attenuation(GO:0009386) |
| 0.9 | 5.5 | GO:0070170 | regulation of tooth mineralization(GO:0070170) |
| 0.9 | 4.5 | GO:0061042 | vascular wound healing(GO:0061042) |
| 0.9 | 4.5 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.9 | 2.7 | GO:0021718 | superior olivary nucleus development(GO:0021718) superior olivary nucleus maturation(GO:0021722) |
| 0.9 | 3.5 | GO:0042539 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.9 | 3.5 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.9 | 3.4 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.8 | 4.2 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.8 | 3.3 | GO:0005986 | sucrose biosynthetic process(GO:0005986) |
| 0.8 | 4.9 | GO:0010909 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.8 | 10.5 | GO:0002467 | germinal center formation(GO:0002467) |
| 0.8 | 3.2 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.8 | 1.6 | GO:0060061 | Spemann organizer formation(GO:0060061) canonical Wnt signaling pathway involved in midbrain dopaminergic neuron differentiation(GO:1904954) |
| 0.8 | 3.1 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.8 | 2.3 | GO:1903033 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.8 | 10.5 | GO:2000394 | positive regulation of lamellipodium morphogenesis(GO:2000394) |
| 0.8 | 13.5 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.7 | 1.5 | GO:0090035 | regulation of chaperone-mediated protein complex assembly(GO:0090034) positive regulation of chaperone-mediated protein complex assembly(GO:0090035) |
| 0.7 | 5.1 | GO:0070305 | response to cGMP(GO:0070305) |
| 0.7 | 5.0 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.7 | 2.1 | GO:0048867 | stem cell fate determination(GO:0048867) |
| 0.7 | 2.8 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.7 | 1.4 | GO:0060298 | positive regulation of sarcomere organization(GO:0060298) |
| 0.7 | 7.4 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.7 | 35.3 | GO:0031529 | ruffle organization(GO:0031529) |
| 0.6 | 1.9 | GO:2000296 | negative regulation of hydrogen peroxide catabolic process(GO:2000296) |
| 0.6 | 1.9 | GO:0016095 | polyprenol catabolic process(GO:0016095) |
| 0.6 | 2.5 | GO:0019087 | transformation of host cell by virus(GO:0019087) |
| 0.6 | 8.1 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.6 | 5.6 | GO:0060355 | positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.6 | 7.4 | GO:1904706 | negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 0.6 | 2.5 | GO:0021997 | response to chlorate(GO:0010157) neural plate axis specification(GO:0021997) |
| 0.6 | 2.4 | GO:1905229 | cellular response to glycoprotein(GO:1904588) cellular response to thyrotropin-releasing hormone(GO:1905229) |
| 0.6 | 2.4 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.6 | 3.5 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.6 | 5.8 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.6 | 2.3 | GO:1902723 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.6 | 2.3 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.6 | 2.9 | GO:0042095 | interferon-gamma biosynthetic process(GO:0042095) regulation of interferon-gamma biosynthetic process(GO:0045072) positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.6 | 2.3 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.6 | 1.1 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.6 | 6.2 | GO:2000580 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.6 | 4.5 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.6 | 2.8 | GO:1904073 | trophectodermal cell proliferation(GO:0001834) regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.5 | 2.2 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.5 | 2.7 | GO:0002725 | negative regulation of T cell cytokine production(GO:0002725) |
| 0.5 | 5.9 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.5 | 3.2 | GO:2000330 | positive regulation of T-helper 17 cell lineage commitment(GO:2000330) |
| 0.5 | 1.6 | GO:0035993 | subthalamic nucleus development(GO:0021763) deltoid tuberosity development(GO:0035993) prolactin secreting cell differentiation(GO:0060127) left lung morphogenesis(GO:0060460) pulmonary vein morphogenesis(GO:0060577) superior vena cava morphogenesis(GO:0060578) cell proliferation involved in outflow tract morphogenesis(GO:0061325) |
| 0.5 | 4.2 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.5 | 2.6 | GO:0033489 | cholesterol biosynthetic process via desmosterol(GO:0033489) cholesterol biosynthetic process via lathosterol(GO:0033490) |
| 0.5 | 1.6 | GO:0015783 | GDP-fucose transport(GO:0015783) purine nucleotide-sugar transport(GO:0036079) |
| 0.5 | 3.6 | GO:0010269 | response to selenium ion(GO:0010269) |
| 0.5 | 2.0 | GO:0032431 | activation of phospholipase A2 activity(GO:0032431) |
| 0.5 | 1.5 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.5 | 2.5 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.5 | 2.5 | GO:0089712 | L-aspartate transport(GO:0070778) L-aspartate transmembrane transport(GO:0089712) |
| 0.5 | 4.9 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.5 | 4.9 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.5 | 4.8 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.5 | 1.4 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.5 | 2.4 | GO:0006041 | glucosamine metabolic process(GO:0006041) glucose 1-phosphate metabolic process(GO:0019255) |
| 0.5 | 1.9 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.5 | 7.0 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.5 | 3.2 | GO:0016102 | retinoic acid biosynthetic process(GO:0002138) diterpenoid biosynthetic process(GO:0016102) |
| 0.5 | 4.6 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.4 | 1.3 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.4 | 1.3 | GO:0003413 | chondrocyte differentiation involved in endochondral bone morphogenesis(GO:0003413) |
| 0.4 | 1.7 | GO:2000342 | negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
| 0.4 | 3.0 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.4 | 2.5 | GO:0086042 | cardiac muscle cell-cardiac muscle cell adhesion(GO:0086042) bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.4 | 2.5 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.4 | 0.8 | GO:0035585 | calcium-mediated signaling using extracellular calcium source(GO:0035585) |
| 0.4 | 3.2 | GO:0060689 | cell differentiation involved in salivary gland development(GO:0060689) |
| 0.4 | 1.2 | GO:0071288 | cellular response to mercury ion(GO:0071288) |
| 0.4 | 11.2 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.4 | 1.2 | GO:1902617 | response to fluoride(GO:1902617) |
| 0.4 | 65.0 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.4 | 28.1 | GO:0030049 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.4 | 3.2 | GO:1900227 | positive regulation of NLRP3 inflammasome complex assembly(GO:1900227) |
| 0.4 | 4.3 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.4 | 2.3 | GO:0060174 | limb bud formation(GO:0060174) |
| 0.4 | 1.5 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.4 | 1.1 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.4 | 8.7 | GO:0045601 | regulation of endothelial cell differentiation(GO:0045601) |
| 0.4 | 3.3 | GO:0060750 | epithelial cell proliferation involved in mammary gland duct elongation(GO:0060750) |
| 0.4 | 2.6 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.4 | 4.0 | GO:0048024 | regulation of mRNA splicing, via spliceosome(GO:0048024) |
| 0.4 | 1.4 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.4 | 3.2 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.3 | 5.6 | GO:0008584 | gonad development(GO:0008406) male gonad development(GO:0008584) development of primary male sexual characteristics(GO:0046546) |
| 0.3 | 4.5 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.3 | 4.1 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.3 | 2.0 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.3 | 3.1 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.3 | 2.4 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.3 | 7.5 | GO:0033622 | integrin activation(GO:0033622) |
| 0.3 | 1.3 | GO:0032754 | positive regulation of interleukin-5 production(GO:0032754) |
| 0.3 | 1.0 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.3 | 2.3 | GO:0086048 | membrane depolarization during bundle of His cell action potential(GO:0086048) |
| 0.3 | 2.0 | GO:0002803 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.3 | 0.7 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.3 | 1.0 | GO:0042822 | pyridoxal phosphate metabolic process(GO:0042822) pyridoxal phosphate biosynthetic process(GO:0042823) |
| 0.3 | 2.3 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.3 | 2.9 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.3 | 2.6 | GO:0060449 | bud elongation involved in lung branching(GO:0060449) |
| 0.3 | 3.2 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.3 | 0.3 | GO:1903659 | regulation of complement-dependent cytotoxicity(GO:1903659) |
| 0.3 | 2.2 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.3 | 6.9 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.3 | 1.2 | GO:0072675 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.3 | 1.5 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.3 | 1.2 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.3 | 3.6 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.3 | 3.6 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.3 | 1.2 | GO:0010871 | negative regulation of receptor biosynthetic process(GO:0010871) |
| 0.3 | 1.7 | GO:0052551 | response to defense-related nitric oxide production by other organism involved in symbiotic interaction(GO:0052551) response to defense-related host nitric oxide production(GO:0052565) |
| 0.3 | 12.4 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.3 | 1.4 | GO:0015840 | urea transport(GO:0015840) |
| 0.3 | 1.7 | GO:0014043 | negative regulation of neuron maturation(GO:0014043) |
| 0.3 | 4.3 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.3 | 2.5 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.3 | 2.3 | GO:2000773 | negative regulation of cellular senescence(GO:2000773) |
| 0.3 | 3.5 | GO:0060850 | regulation of transcription involved in cell fate commitment(GO:0060850) |
| 0.3 | 0.5 | GO:0070120 | ciliary neurotrophic factor-mediated signaling pathway(GO:0070120) |
| 0.3 | 1.8 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.3 | 1.3 | GO:0021563 | glossopharyngeal nerve development(GO:0021563) glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.3 | 2.0 | GO:1901374 | acetate ester transport(GO:1901374) |
| 0.3 | 1.0 | GO:0002879 | positive regulation of acute inflammatory response to non-antigenic stimulus(GO:0002879) |
| 0.2 | 1.2 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.2 | 3.5 | GO:1901750 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.2 | 16.3 | GO:0031016 | pancreas development(GO:0031016) |
| 0.2 | 0.7 | GO:0010644 | cell communication by electrical coupling(GO:0010644) |
| 0.2 | 0.7 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.2 | 1.5 | GO:0042364 | water-soluble vitamin biosynthetic process(GO:0042364) |
| 0.2 | 0.7 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.2 | 0.2 | GO:0071638 | negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.2 | 1.4 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.2 | 3.8 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.2 | 0.9 | GO:0051549 | positive regulation of keratinocyte migration(GO:0051549) |
| 0.2 | 1.2 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.2 | 3.4 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.2 | 2.0 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.2 | 1.1 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.2 | 6.3 | GO:0051281 | positive regulation of release of sequestered calcium ion into cytosol(GO:0051281) |
| 0.2 | 6.4 | GO:1901687 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.2 | 0.6 | GO:0045645 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 0.2 | 3.8 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.2 | 1.0 | GO:0008595 | tripartite regional subdivision(GO:0007351) anterior/posterior axis specification, embryo(GO:0008595) |
| 0.2 | 1.7 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.2 | 4.0 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.2 | 1.9 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.2 | 1.5 | GO:0098856 | intestinal cholesterol absorption(GO:0030299) intestinal lipid absorption(GO:0098856) |
| 0.2 | 10.4 | GO:0031102 | neuron projection regeneration(GO:0031102) |
| 0.2 | 1.5 | GO:1904354 | negative regulation of telomere capping(GO:1904354) |
| 0.2 | 2.6 | GO:0060047 | heart process(GO:0003015) heart contraction(GO:0060047) |
| 0.2 | 6.0 | GO:0006693 | prostanoid metabolic process(GO:0006692) prostaglandin metabolic process(GO:0006693) |
| 0.2 | 1.6 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.2 | 0.5 | GO:0060581 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) ventral spinal cord interneuron fate commitment(GO:0060579) cell fate commitment involved in pattern specification(GO:0060581) |
| 0.2 | 1.2 | GO:0051712 | positive regulation of killing of cells of other organism(GO:0051712) |
| 0.2 | 1.7 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.2 | 4.0 | GO:0071711 | basement membrane organization(GO:0071711) |
| 0.2 | 2.3 | GO:0046320 | regulation of fatty acid oxidation(GO:0046320) |
| 0.2 | 1.9 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.2 | 1.5 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.2 | 0.5 | GO:0098935 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) |
| 0.2 | 0.6 | GO:0097688 | AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) |
| 0.2 | 2.0 | GO:0042984 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.2 | 0.9 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.2 | 0.5 | GO:2000653 | regulation of genetic imprinting(GO:2000653) |
| 0.1 | 2.1 | GO:0045954 | positive regulation of natural killer cell mediated cytotoxicity(GO:0045954) |
| 0.1 | 0.9 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.1 | 2.7 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.1 | 6.3 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.1 | 2.2 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.1 | 2.5 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.1 | 3.3 | GO:0050974 | detection of mechanical stimulus involved in sensory perception(GO:0050974) |
| 0.1 | 0.8 | GO:0038042 | dimeric G-protein coupled receptor signaling pathway(GO:0038042) |
| 0.1 | 1.4 | GO:2000020 | positive regulation of male gonad development(GO:2000020) |
| 0.1 | 2.3 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.1 | 7.8 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 6.6 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.1 | 1.1 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.1 | 3.9 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.1 | 1.2 | GO:0010886 | positive regulation of cholesterol storage(GO:0010886) |
| 0.1 | 5.1 | GO:0048706 | embryonic skeletal system development(GO:0048706) |
| 0.1 | 0.7 | GO:0060290 | transforming growth factor beta3 production(GO:0032907) regulation of transforming growth factor beta3 production(GO:0032910) positive regulation of transforming growth factor beta3 production(GO:0032916) transdifferentiation(GO:0060290) negative regulation of lung blood pressure(GO:0061767) |
| 0.1 | 1.1 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.1 | 4.4 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.1 | 1.5 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.1 | 0.2 | GO:1904338 | regulation of dopaminergic neuron differentiation(GO:1904338) |
| 0.1 | 2.5 | GO:0017144 | drug metabolic process(GO:0017144) |
| 0.1 | 7.4 | GO:0061178 | regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.1 | 2.2 | GO:0090190 | positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.1 | 3.1 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.1 | 8.8 | GO:0006027 | glycosaminoglycan catabolic process(GO:0006027) |
| 0.1 | 1.0 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.1 | 3.0 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.1 | 1.7 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 1.3 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.1 | 2.0 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.1 | 8.9 | GO:0002062 | chondrocyte differentiation(GO:0002062) |
| 0.1 | 3.5 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.1 | 3.3 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.1 | 3.4 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.1 | 0.8 | GO:0099612 | protein localization to axon(GO:0099612) |
| 0.1 | 1.4 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.1 | 0.5 | GO:1904885 | beta-catenin destruction complex assembly(GO:1904885) |
| 0.1 | 3.6 | GO:0046710 | GDP metabolic process(GO:0046710) |
| 0.1 | 1.3 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.1 | 4.3 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.1 | 1.9 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.1 | 5.9 | GO:0000271 | polysaccharide biosynthetic process(GO:0000271) |
| 0.1 | 0.4 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.1 | 3.5 | GO:0008038 | neuron recognition(GO:0008038) |
| 0.1 | 1.3 | GO:0045104 | intermediate filament cytoskeleton organization(GO:0045104) |
| 0.1 | 0.8 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.1 | 7.0 | GO:0032508 | DNA duplex unwinding(GO:0032508) |
| 0.1 | 3.0 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.1 | 3.3 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.1 | 2.7 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.1 | 1.1 | GO:1901673 | regulation of mitotic spindle assembly(GO:1901673) |
| 0.1 | 0.4 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.1 | 1.1 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.1 | 1.0 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.1 | 2.1 | GO:0007129 | synapsis(GO:0007129) |
| 0.1 | 0.9 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.1 | 3.6 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.1 | 7.2 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.1 | 3.3 | GO:0007368 | determination of left/right symmetry(GO:0007368) |
| 0.1 | 1.2 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.1 | 0.7 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.1 | 2.3 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.1 | 0.6 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.1 | 1.3 | GO:0014059 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.1 | 3.7 | GO:0006024 | aminoglycan biosynthetic process(GO:0006023) glycosaminoglycan biosynthetic process(GO:0006024) |
| 0.1 | 1.6 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.1 | 1.8 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.1 | 2.7 | GO:0006970 | response to osmotic stress(GO:0006970) |
| 0.1 | 1.8 | GO:1901185 | negative regulation of ERBB signaling pathway(GO:1901185) |
| 0.1 | 0.3 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.1 | 0.6 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.1 | 2.2 | GO:0042472 | inner ear morphogenesis(GO:0042472) |
| 0.0 | 1.8 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 1.8 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.2 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.1 | GO:0097152 | mesenchymal cell apoptotic process(GO:0097152) |
| 0.0 | 1.2 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 2.0 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.4 | GO:0060259 | regulation of feeding behavior(GO:0060259) |
| 0.0 | 0.1 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 0.0 | 1.4 | GO:0046949 | fatty-acyl-CoA biosynthetic process(GO:0046949) |
| 0.0 | 2.8 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) |
| 0.0 | 0.5 | GO:0031424 | keratinization(GO:0031424) |
| 0.0 | 0.3 | GO:0032930 | positive regulation of superoxide anion generation(GO:0032930) |
| 0.0 | 0.2 | GO:0048711 | positive regulation of astrocyte differentiation(GO:0048711) |
| 0.0 | 2.2 | GO:1990748 | cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.0 | 0.7 | GO:0045332 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.0 | 1.2 | GO:0048705 | skeletal system morphogenesis(GO:0048705) |
| 0.0 | 0.2 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.6 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.3 | GO:0021522 | spinal cord motor neuron differentiation(GO:0021522) |
| 0.0 | 1.4 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 1.2 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.0 | 2.3 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.0 | 0.3 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.5 | GO:0043542 | endothelial cell migration(GO:0043542) |
| 0.0 | 0.1 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.0 | 0.7 | GO:0060976 | coronary vasculature development(GO:0060976) |
| 0.0 | 1.1 | GO:0051216 | cartilage development(GO:0051216) |
| 0.0 | 1.1 | GO:0045995 | regulation of embryonic development(GO:0045995) |
| 0.0 | 0.1 | GO:0006581 | acetylcholine catabolic process in synaptic cleft(GO:0001507) acetylcholine catabolic process(GO:0006581) negative regulation of synaptic transmission, cholinergic(GO:0032223) neurotransmitter receptor biosynthetic process(GO:0045212) neurotransmitter receptor metabolic process(GO:0045213) |
| 0.0 | 1.7 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 1.1 | GO:0007498 | mesoderm development(GO:0007498) |
| 0.0 | 0.6 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 1.3 | GO:0050796 | regulation of insulin secretion(GO:0050796) |
| 0.0 | 1.5 | GO:0014065 | phosphatidylinositol 3-kinase signaling(GO:0014065) |
| 0.0 | 3.6 | GO:0051056 | regulation of small GTPase mediated signal transduction(GO:0051056) |
| 0.0 | 1.6 | GO:0002576 | platelet degranulation(GO:0002576) |
| 0.0 | 1.0 | GO:0070268 | cornification(GO:0070268) |
| 0.0 | 0.4 | GO:0001578 | microtubule bundle formation(GO:0001578) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 16.2 | 226.8 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 6.4 | 19.3 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 4.4 | 44.3 | GO:0071953 | elastic fiber(GO:0071953) |
| 4.3 | 17.2 | GO:0045160 | myosin I complex(GO:0045160) |
| 4.1 | 77.2 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 2.7 | 29.6 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 1.6 | 24.5 | GO:0030478 | actin cap(GO:0030478) |
| 1.6 | 11.4 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 1.5 | 5.8 | GO:1990246 | uniplex complex(GO:1990246) |
| 1.3 | 3.8 | GO:0097489 | multivesicular body, internal vesicle lumen(GO:0097489) |
| 1.3 | 6.3 | GO:0060201 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 1.1 | 4.6 | GO:0005600 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
| 1.1 | 14.6 | GO:0097433 | dense body(GO:0097433) |
| 0.9 | 4.3 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.8 | 13.5 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.8 | 13.4 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.8 | 11.6 | GO:0008091 | spectrin(GO:0008091) |
| 0.8 | 4.9 | GO:1990130 | EGO complex(GO:0034448) Iml1 complex(GO:1990130) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.8 | 4.9 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.8 | 6.1 | GO:0005593 | FACIT collagen trimer(GO:0005593) |
| 0.7 | 4.9 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.7 | 1.4 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.7 | 7.9 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.6 | 4.5 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.6 | 44.4 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.6 | 2.4 | GO:0008043 | intracellular ferritin complex(GO:0008043) ferritin complex(GO:0070288) |
| 0.6 | 2.3 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.6 | 2.2 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.5 | 6.0 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.5 | 4.9 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.5 | 2.6 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.5 | 18.1 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.5 | 1.9 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.5 | 1.9 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 0.5 | 3.2 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.4 | 1.3 | GO:0043159 | perinuclear theca(GO:0033011) cytoskeletal calyx(GO:0033150) acrosomal matrix(GO:0043159) |
| 0.4 | 12.9 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.4 | 2.3 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.4 | 11.7 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.4 | 43.5 | GO:0005604 | basement membrane(GO:0005604) |
| 0.4 | 1.5 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.4 | 1.4 | GO:0030430 | host cell cytoplasm(GO:0030430) host cell cytoplasm part(GO:0033655) |
| 0.3 | 19.2 | GO:0016235 | aggresome(GO:0016235) |
| 0.3 | 2.6 | GO:0016012 | dystroglycan complex(GO:0016011) sarcoglycan complex(GO:0016012) |
| 0.3 | 8.9 | GO:0031312 | extrinsic component of organelle membrane(GO:0031312) |
| 0.3 | 2.5 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.3 | 3.3 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.3 | 4.6 | GO:0005883 | neurofilament(GO:0005883) |
| 0.3 | 5.7 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.3 | 2.3 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.3 | 2.0 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.3 | 8.3 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.3 | 7.9 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.3 | 7.3 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.3 | 0.5 | GO:0070110 | ciliary neurotrophic factor receptor complex(GO:0070110) |
| 0.3 | 27.8 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.3 | 1.5 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.3 | 2.5 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.2 | 3.4 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.2 | 3.1 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.2 | 6.5 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.2 | 0.2 | GO:0097059 | CNTFR-CLCF1 complex(GO:0097059) |
| 0.2 | 1.4 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.2 | 61.2 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.2 | 9.7 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.2 | 0.8 | GO:0038038 | G-protein coupled receptor homodimeric complex(GO:0038038) |
| 0.2 | 5.0 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.2 | 4.1 | GO:0030286 | dynein complex(GO:0030286) |
| 0.2 | 0.6 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.2 | 7.4 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.2 | 4.8 | GO:1990752 | microtubule end(GO:1990752) |
| 0.2 | 11.8 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.2 | 13.5 | GO:0005811 | lipid particle(GO:0005811) |
| 0.2 | 12.7 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.2 | 3.9 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.2 | 10.2 | GO:0005776 | autophagosome(GO:0005776) |
| 0.1 | 11.6 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 2.0 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.1 | 2.6 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.1 | 12.5 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 3.3 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 1.4 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 14.3 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 0.7 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.1 | 11.6 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.1 | 4.2 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 16.5 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.1 | 7.9 | GO:0005902 | microvillus(GO:0005902) |
| 0.1 | 2.3 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 0.5 | GO:0031310 | intrinsic component of endosome membrane(GO:0031302) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.1 | 12.8 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 3.4 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.1 | 1.1 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 19.7 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 0.3 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.1 | 2.0 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 4.2 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.1 | 0.5 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 1.3 | GO:0043073 | germ cell nucleus(GO:0043073) |
| 0.1 | 0.9 | GO:0032982 | myosin filament(GO:0032982) |
| 0.1 | 0.7 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 2.3 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.1 | 8.9 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.1 | 11.7 | GO:0030018 | Z disc(GO:0030018) |
| 0.1 | 11.3 | GO:0005938 | cell cortex(GO:0005938) |
| 0.1 | 0.6 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.1 | 0.5 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.1 | 18.6 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.1 | 2.9 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 0.4 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.1 | 0.2 | GO:0045179 | apical cortex(GO:0045179) |
| 0.1 | 4.9 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 0.7 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 2.5 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.1 | 0.4 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 0.7 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 4.2 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 3.5 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.1 | 1.5 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.1 | 1.8 | GO:0044291 | cell-cell contact zone(GO:0044291) |
| 0.1 | 10.3 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 0.9 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 1.4 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.0 | 0.6 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 65.6 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 1.2 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 1.5 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 1.5 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 0.7 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.8 | 23.3 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 3.8 | 11.5 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 3.6 | 10.9 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 2.7 | 197.4 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 2.2 | 6.7 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 2.2 | 6.7 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 2.2 | 6.5 | GO:0070546 | L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 2.1 | 12.9 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 2.0 | 14.2 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 1.9 | 9.5 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 1.9 | 7.6 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 1.9 | 7.5 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 1.8 | 7.1 | GO:0097001 | ceramide binding(GO:0097001) |
| 1.7 | 6.6 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 1.6 | 9.7 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 1.6 | 8.0 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 1.6 | 4.7 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 1.5 | 1.5 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 1.5 | 29.6 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 1.4 | 5.7 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 1.4 | 10.9 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 1.4 | 4.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 1.3 | 6.3 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 1.2 | 8.7 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 1.2 | 11.6 | GO:0042731 | PH domain binding(GO:0042731) |
| 1.2 | 3.5 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 1.1 | 13.6 | GO:0045159 | myosin II binding(GO:0045159) |
| 1.1 | 4.5 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 1.1 | 3.4 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 1.1 | 3.3 | GO:0098640 | integrin binding involved in cell-matrix adhesion(GO:0098640) |
| 1.1 | 11.8 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 1.0 | 4.0 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.9 | 91.2 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.9 | 7.5 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.9 | 5.5 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.9 | 97.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.9 | 31.9 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.9 | 2.7 | GO:0016534 | cyclin-dependent protein kinase 5 activator activity(GO:0016534) |
| 0.9 | 3.5 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.8 | 3.4 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.8 | 10.8 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.8 | 3.3 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 0.8 | 12.8 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.8 | 2.4 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.8 | 3.1 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.8 | 3.0 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.8 | 1.5 | GO:0019828 | aspartic-type endopeptidase inhibitor activity(GO:0019828) |
| 0.8 | 3.8 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.7 | 1.5 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.7 | 5.9 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.7 | 9.6 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.7 | 15.6 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.7 | 28.4 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.7 | 13.1 | GO:0008392 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.7 | 2.0 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.7 | 3.3 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.6 | 1.9 | GO:0003865 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.6 | 2.5 | GO:0033906 | protein tyrosine kinase inhibitor activity(GO:0030292) hyaluronoglucuronidase activity(GO:0033906) |
| 0.6 | 3.2 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.6 | 6.9 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.6 | 33.0 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.6 | 3.7 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.6 | 3.0 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.6 | 1.2 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.6 | 2.3 | GO:0035473 | lipase binding(GO:0035473) |
| 0.6 | 4.6 | GO:0005534 | galactose binding(GO:0005534) |
| 0.6 | 14.6 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.5 | 3.3 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.5 | 10.7 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.5 | 3.7 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.5 | 3.2 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.5 | 10.6 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.5 | 1.6 | GO:0005457 | GDP-fucose transmembrane transporter activity(GO:0005457) purine nucleotide-sugar transmembrane transporter activity(GO:0036080) |
| 0.5 | 13.2 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.5 | 15.7 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.5 | 3.5 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.5 | 2.0 | GO:1901375 | acetylcholine transmembrane transporter activity(GO:0005277) secondary active organic cation transmembrane transporter activity(GO:0008513) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.5 | 4.5 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.5 | 2.5 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.5 | 7.0 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.5 | 2.5 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.5 | 2.5 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.5 | 1.9 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.5 | 6.7 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.5 | 3.3 | GO:0001093 | TFIIB-class transcription factor binding(GO:0001093) |
| 0.5 | 6.4 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.5 | 6.4 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.4 | 4.4 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.4 | 1.3 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.4 | 3.9 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.4 | 6.7 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.4 | 5.0 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.4 | 2.5 | GO:0015185 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 0.4 | 4.4 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.4 | 2.4 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.4 | 2.3 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.4 | 5.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.4 | 2.6 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.4 | 4.8 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.4 | 1.5 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.4 | 7.5 | GO:0005112 | Notch binding(GO:0005112) |
| 0.3 | 3.4 | GO:0003720 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.3 | 2.4 | GO:0004996 | thyroid-stimulating hormone receptor activity(GO:0004996) |
| 0.3 | 3.4 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.3 | 2.0 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.3 | 2.7 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.3 | 1.0 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.3 | 1.9 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.3 | 1.2 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.3 | 7.2 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.3 | 24.9 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.3 | 1.2 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.3 | 2.0 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.3 | 3.2 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.3 | 1.5 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.3 | 0.9 | GO:0016215 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.3 | 3.1 | GO:0086007 | voltage-gated calcium channel activity involved in cardiac muscle cell action potential(GO:0086007) |
| 0.3 | 1.4 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.3 | 5.4 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.3 | 1.7 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.3 | 1.4 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.3 | 3.4 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.3 | 2.5 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.3 | 2.2 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.3 | 5.5 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.3 | 7.9 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.3 | 5.1 | GO:0005537 | mannose binding(GO:0005537) |
| 0.3 | 12.0 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.3 | 2.4 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.3 | 1.3 | GO:0005113 | patched binding(GO:0005113) |
| 0.3 | 0.5 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.3 | 1.8 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.3 | 1.3 | GO:0043813 | phosphatidylinositol-3,5-bisphosphate 5-phosphatase activity(GO:0043813) |
| 0.3 | 9.2 | GO:0005518 | collagen binding(GO:0005518) |
| 0.3 | 36.0 | GO:0005178 | integrin binding(GO:0005178) |
| 0.2 | 7.2 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.2 | 3.5 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.2 | 7.2 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.2 | 8.6 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.2 | 1.7 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.2 | 4.3 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.2 | 0.7 | GO:0000035 | acyl binding(GO:0000035) |
| 0.2 | 2.4 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.2 | 1.0 | GO:0032558 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.2 | 11.9 | GO:0032934 | cholesterol binding(GO:0015485) sterol binding(GO:0032934) |
| 0.2 | 2.3 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.2 | 5.7 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.2 | 35.1 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.2 | 0.6 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.2 | 0.6 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 0.2 | 0.6 | GO:0031812 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.2 | 2.5 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.2 | 7.2 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.2 | 3.9 | GO:0042166 | acetylcholine binding(GO:0042166) |
| 0.2 | 6.7 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.2 | 2.1 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.2 | 1.2 | GO:0015168 | glycerol transmembrane transporter activity(GO:0015168) |
| 0.2 | 2.0 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.2 | 2.5 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.2 | 1.0 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.2 | 3.4 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.2 | 8.3 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.2 | 1.5 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 11.1 | GO:0001190 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.1 | 3.2 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 0.7 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.1 | 2.3 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 3.9 | GO:0001848 | complement binding(GO:0001848) |
| 0.1 | 1.5 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 2.2 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.1 | 6.4 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 0.9 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.1 | 2.5 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.1 | 0.6 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.1 | 0.2 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.1 | 8.0 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.1 | 0.3 | GO:0017082 | mineralocorticoid receptor activity(GO:0017082) |
| 0.1 | 1.7 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.1 | 3.1 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 0.4 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.1 | 0.7 | GO:1903763 | gap junction channel activity involved in cell communication by electrical coupling(GO:1903763) |
| 0.1 | 3.2 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.1 | 0.8 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.1 | 2.9 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 1.1 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.1 | 5.3 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 2.2 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.1 | 0.9 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 8.3 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 10.5 | GO:0005179 | hormone activity(GO:0005179) |
| 0.1 | 1.7 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 1.7 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.1 | 1.0 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 0.7 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.1 | 1.0 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.1 | 0.9 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.1 | 1.5 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.1 | 2.5 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.1 | 2.6 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 2.6 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.1 | 1.2 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 5.6 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 1.7 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 1.0 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.1 | 0.5 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.1 | 1.7 | GO:0002020 | protease binding(GO:0002020) |
| 0.1 | 3.1 | GO:0070851 | growth factor receptor binding(GO:0070851) |
| 0.1 | 1.5 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.1 | 5.0 | GO:0008201 | heparin binding(GO:0008201) |
| 0.1 | 1.7 | GO:0001191 | transcriptional repressor activity, RNA polymerase II transcription factor binding(GO:0001191) |
| 0.1 | 10.6 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.1 | 2.5 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.4 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 1.3 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 2.0 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) |
| 0.0 | 0.2 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.6 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.0 | 1.5 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 9.7 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 1.7 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 0.7 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.0 | 3.9 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 2.0 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 1.8 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 0.6 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 5.8 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.1 | GO:0016453 | C-acetyltransferase activity(GO:0016453) |
| 0.0 | 0.6 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.6 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 1.1 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.7 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.0 | 0.7 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.6 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 1.0 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.3 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 8.6 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 0.5 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.2 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.5 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 1.7 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 1.2 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 161.9 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 2.3 | 109.2 | NABA COLLAGENS | Genes encoding collagen proteins |
| 1.1 | 71.0 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.6 | 5.9 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.5 | 2.2 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.5 | 2.7 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.4 | 10.7 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.4 | 79.2 | NABA CORE MATRISOME | Ensemble of genes encoding core extracellular matrix including ECM glycoproteins, collagens and proteoglycans |
| 0.4 | 26.3 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.4 | 14.0 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.3 | 11.8 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.3 | 14.0 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.3 | 9.8 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.3 | 10.3 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.3 | 3.2 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.3 | 1.6 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.3 | 6.4 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.2 | 2.3 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.2 | 3.8 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.2 | 58.0 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.2 | 6.8 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.2 | 9.4 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.2 | 14.5 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.2 | 5.3 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.2 | 8.2 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.2 | 7.8 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.2 | 7.7 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.2 | 7.9 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.2 | 8.6 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.2 | 9.4 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.2 | 3.1 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.2 | 3.8 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.2 | 9.5 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.2 | 14.6 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 3.7 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.1 | 4.5 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 5.7 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 9.1 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.1 | 2.2 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 2.4 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.1 | 3.8 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 4.7 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.1 | 16.9 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 6.2 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.1 | 2.3 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 3.5 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.1 | 5.7 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 1.6 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.1 | 4.5 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.1 | 1.2 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 1.1 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.1 | 1.0 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.1 | 1.3 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 1.2 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.1 | 1.0 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 1.6 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 0.3 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.3 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 2.2 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.4 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.9 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 1.0 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.5 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 1.4 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.8 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.5 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.9 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.3 | 142.0 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 1.8 | 119.0 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 1.3 | 85.0 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 1.3 | 1.3 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 1.2 | 23.3 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 1.1 | 11.8 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.7 | 13.1 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.7 | 9.2 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.6 | 12.3 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.6 | 15.0 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.5 | 17.5 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.5 | 8.3 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.5 | 2.3 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.4 | 4.9 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.4 | 14.2 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.4 | 8.9 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.4 | 3.2 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.3 | 5.5 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.3 | 4.4 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.3 | 4.3 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.3 | 6.8 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.3 | 27.6 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.3 | 6.4 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.3 | 6.5 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.3 | 3.8 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.3 | 6.7 | REACTOME ACYL CHAIN REMODELLING OF PC | Genes involved in Acyl chain remodelling of PC |
| 0.3 | 4.8 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.2 | 6.9 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.2 | 5.6 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.2 | 4.8 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.2 | 4.3 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.2 | 3.9 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.2 | 7.8 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.2 | 3.4 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.2 | 6.8 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.2 | 2.5 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.2 | 10.0 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.2 | 7.3 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.2 | 2.1 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.2 | 6.4 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.2 | 6.2 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.2 | 1.8 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.2 | 2.6 | REACTOME INCRETIN SYNTHESIS SECRETION AND INACTIVATION | Genes involved in Incretin Synthesis, Secretion, and Inactivation |
| 0.2 | 4.9 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.2 | 4.9 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.2 | 2.3 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.1 | 2.4 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 1.9 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.1 | 3.0 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 2.2 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.1 | 2.7 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.1 | 0.9 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 2.4 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 1.8 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.1 | 9.7 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 3.4 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 2.0 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.1 | 4.6 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 2.7 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.1 | 2.3 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.1 | 2.9 | REACTOME SHC MEDIATED CASCADE | Genes involved in SHC-mediated cascade |
| 0.1 | 1.7 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 1.8 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 1.7 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 1.9 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.1 | 3.0 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.1 | 1.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 1.1 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.1 | 2.4 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 13.6 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 4.2 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.1 | 1.3 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.1 | 1.2 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 7.8 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.1 | 0.7 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.1 | 1.7 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.1 | 0.9 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 0.6 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.1 | 2.4 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.1 | 2.1 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.1 | 1.6 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.1 | 1.9 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 1.6 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 2.4 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 1.0 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.3 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 9.3 | REACTOME GASTRIN CREB SIGNALLING PATHWAY VIA PKC AND MAPK | Genes involved in Gastrin-CREB signalling pathway via PKC and MAPK |
| 0.0 | 2.0 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.6 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 3.9 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.3 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.4 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.0 | 0.7 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 2.0 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 1.5 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.9 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.3 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.0 | 1.1 | REACTOME INTEGRATION OF ENERGY METABOLISM | Genes involved in Integration of energy metabolism |
| 0.0 | 0.5 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |