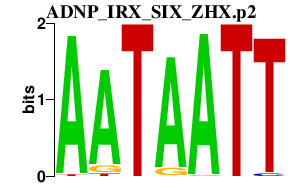
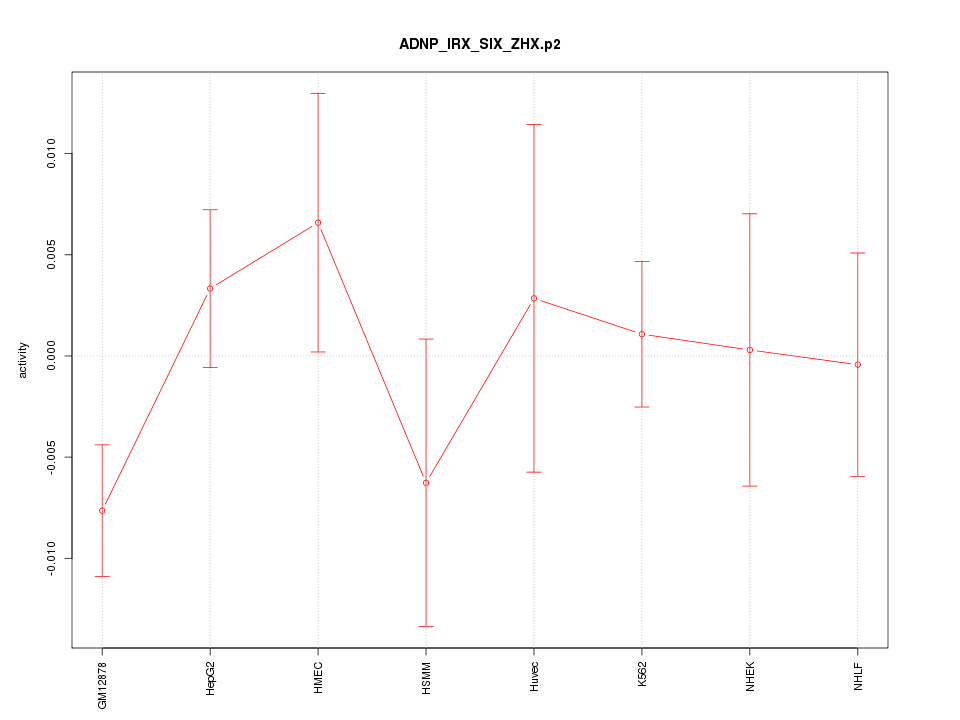
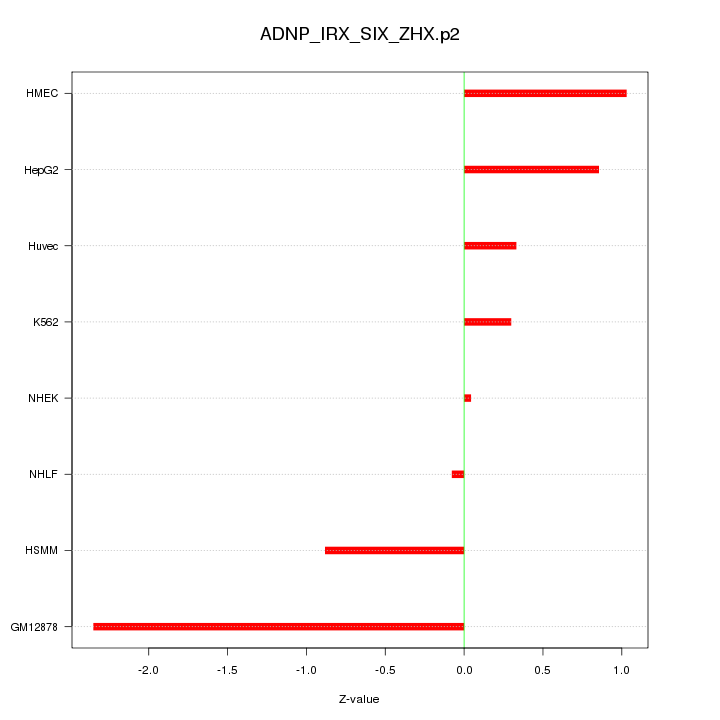
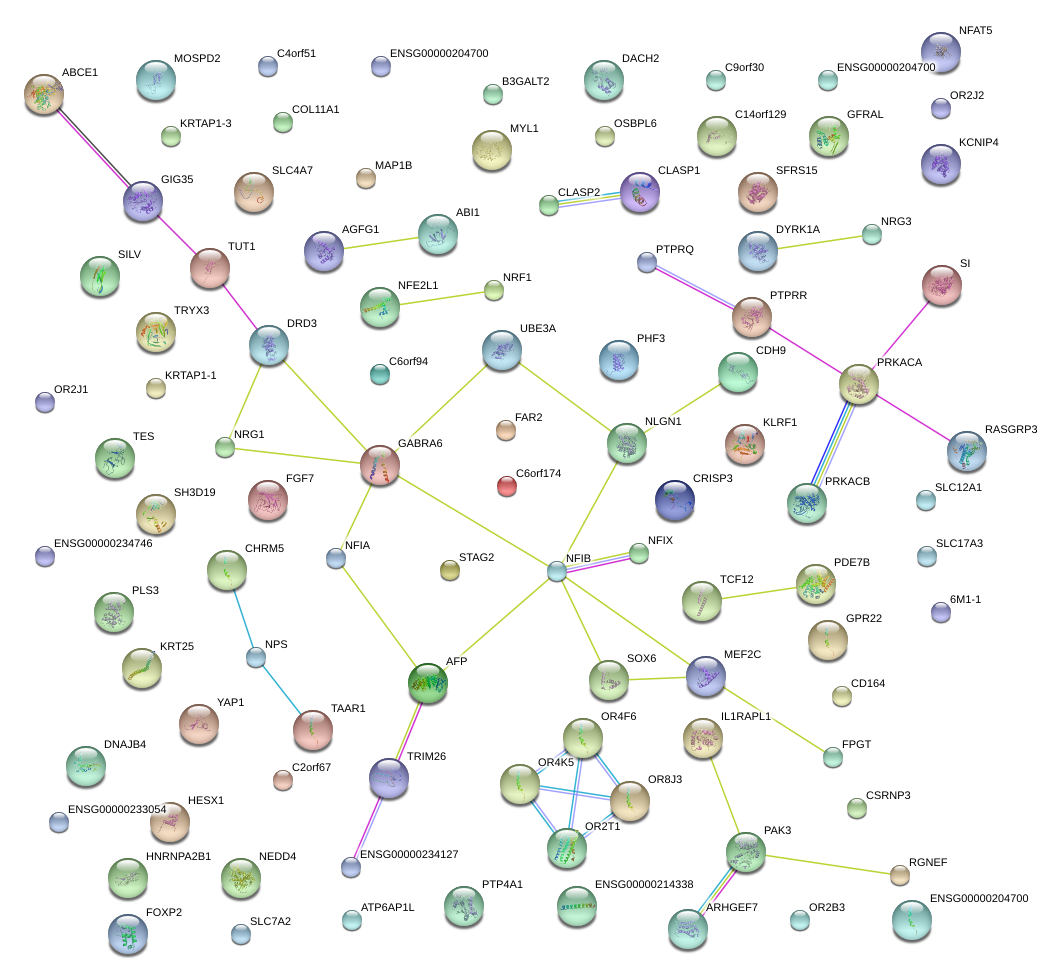
|
chr6_+_64414389
|
3.739
|
NM_015153
|
PHF3
|
PHD finger protein 3
|
|
chr7_-_132371208
|
2.298
|
|
EEF1G
|
eukaryotic translation elongation factor 1 gamma
|
|
chr3_-_33675484
|
1.973
|
|
CLASP2
|
cytoplasmic linker associated protein 2
|
|
chr10_+_129237602
|
1.733
|
NM_001030013
|
NPS
|
neuropeptide S
|
|
chr11_+_101488380
|
1.678
|
NM_001195045
|
YAP1
|
Yes-associated protein 1
|
|
chr9_-_14076901
|
1.672
|
|
NFIB
|
nuclear factor I/B
|
|
chr1_+_84402962
|
1.647
|
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr3_+_174598937
|
1.640
|
NM_014932
|
NLGN1
|
neuroligin 1
|
|
chrX_+_85404626
|
1.627
|
NM_001139515
|
DACH2
|
dachshund homolog 2 (Drosophila)
|
|
chr10_+_83627422
|
1.603
|
NM_001165973
|
NRG3
|
neuregulin 3
|
|
chr6_+_64344307
|
1.602
|
|
PTP4A1
|
protein tyrosine phosphatase type IVA, member 1
|
|
chr4_+_146250978
|
1.598
|
|
ABCE1
|
ATP-binding cassette, sub-family E (OABP), member 1
|
|
chr2_+_166137091
|
1.548
|
NM_024969
|
CSRNP3
|
cysteine-serine-rich nuclear protein 3
|
|
chr3_-_115380539
|
1.537
|
NM_000796
NM_033663
|
DRD3
|
dopamine receptor D3
|
|
chr9_+_102244005
|
1.516
|
NM_001198812
|
C9orf3-TMEFF1
C9orf30
|
C9orf3-TMEFF1 readthrough
chromosome 9 open reading frame 30
|
|
chrX_+_56772391
|
1.430
|
|
LOC550643
|
hypothetical LOC550643
|
|
chr4_+_74520796
|
1.424
|
NM_001134
|
AFP
|
alpha-fetoprotein
|
|
chr7_+_106897737
|
1.373
|
NM_005295
|
GPR22
|
G protein-coupled receptor 22
|
|
chr6_+_29249289
|
1.368
|
NM_030905
|
OR2J2
|
olfactory receptor, family 2, subfamily J, member 2
|
|
chrX_+_114734074
|
1.356
|
NM_001136025
NM_001172335
|
PLS3
|
plastin 3
|
|
chr15_+_47502666
|
1.341
|
NM_002009
|
FGF7
|
fibroblast growth factor 7
|
|
chr7_-_141604323
|
1.325
|
NM_001001317
|
TRYX3
|
trypsin X3
|
|
chr6_-_127882192
|
1.264
|
NM_001012279
|
C6orf174
|
chromosome 6 open reading frame 174
|
|
chrX_+_28515479
|
1.258
|
NM_014271
|
IL1RAPL1
|
interleukin 1 receptor accessory protein-like 1
|
|
chr6_+_144227265
|
1.237
|
NM_001013623
|
C6orf94
|
chromosome 6 open reading frame 94
|
|
chr2_-_210726539
|
1.205
|
|
C2orf67
|
chromosome 2 open reading frame 67
|
|
chr12_+_29267863
|
1.193
|
NM_018099
|
FAR2
|
fatty acyl CoA reductase 2
|
|
chr6_-_29163027
|
1.190
|
NM_001005226
|
OR2B3
|
olfactory receptor, family 2, subfamily B, member 3
|
|
chr3_-_27473248
|
1.116
|
NM_003615
|
SLC4A7
|
solute carrier family 4, sodium bicarbonate cotransporter, member 7
|
|
chr7_+_129057152
|
1.098
|
NM_001040110
|
NRF1
|
nuclear respiratory factor 1
|
|
chr14_+_19458580
|
1.089
|
NM_001005483
|
OR4K5
|
olfactory receptor, family 4, subfamily K, member 5
|
|
chr21_+_37713076
|
1.082
|
NM_130436
|
DYRK1A
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A
|
|
chr3_-_166278969
|
1.074
|
NM_001041
|
SI
|
sucrase-isomaltase (alpha-glucosidase)
|
|
chr6_+_55300225
|
1.064
|
NM_207410
|
GFRAL
|
GDNF family receptor alpha like
|
|
chr7_+_115650240
|
1.060
|
NM_152829
|
TES
|
testis derived transcript (3 LIM domains)
|
|
chr4_-_79322295
|
1.053
|
|
|
|
|
chr8_+_17440664
|
1.008
|
NM_001164771
NM_003046
|
SLC7A2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2
|
|
chr1_+_78243181
|
0.994
|
NM_007034
|
DNAJB4
|
DnaJ (Hsp40) homolog, subfamily B, member 4
|
|
chr5_-_27074407
|
0.976
|
NM_016279
|
CDH9
|
cadherin 9, type 2 (T1-cadherin)
|
|
chr11_-_55661769
|
0.963
|
NM_001004064
|
OR8J3
|
olfactory receptor, family 8, subfamily J, member 3
|
|
chr1_+_74473672
|
0.955
|
NM_015978
|
TNNI3K
|
TNNI3 interacting kinase
|
|
chr3_-_57209319
|
0.954
|
NM_003865
|
HESX1
|
HESX homeobox 1
|
|
chr14_+_95915774
|
0.934
|
NM_016472
|
C14orf129
|
chromosome 14 open reading frame 129
|
|
chr16_+_68238460
|
0.918
|
|
NFAT5
|
nuclear factor of activated T-cells 5, tonicity-responsive
|
|
chr5_+_81636921
|
0.904
|
NM_001017971
|
ATP6AP1L
|
ATPase, H+ transporting, lysosomal accessory protein 1-like
|
|
chr6_-_25982418
|
0.904
|
NM_001098486
NM_006632
|
SLC17A3
|
solute carrier family 17 (sodium phosphate), member 3
|
|
chr2_-_210888010
|
0.887
|
NM_079420
|
MYL1
|
myosin, light chain 1, alkali; skeletal, fast
|
|
chr21_+_24722913
|
0.887
|
|
FLJ42200
|
FLJ42200 protein
|
|
chr5_+_161045235
|
0.884
|
NM_000811
|
GABRA6
|
gamma-aminobutyric acid (GABA) A receptor, alpha 6
|
|
chr15_+_100163424
|
0.880
|
NM_001005326
|
OR4F6
|
olfactory receptor, family 4, subfamily F, member 6
|
|
chr4_+_146820805
|
0.879
|
NM_001080531
|
C4orf51
|
chromosome 4 open reading frame 51
|
|
chr12_+_79362256
|
0.873
|
NM_001145026
|
PTPRQ
|
protein tyrosine phosphatase, receptor type, Q
|
|
chr2_-_121875610
|
0.871
|
|
CLASP1
|
cytoplasmic linker associated protein 1
|
|
chr12_+_9871343
|
0.851
|
NM_016523
|
KLRF1
|
killer cell lectin-like receptor subfamily F, member 1
|
|
chr5_-_168204430
|
0.847
|
|
|
|
|
chr4_-_152366970
|
0.845
|
NM_001009555
NM_001128923
|
SH3D19
|
SH3 domain containing 19
|
|
chr2_+_228064531
|
0.836
|
|
AGFG1
|
ArfGAP with FG repeats 1
|
|
chr5_-_88235624
|
0.836
|
NM_001131005
NM_001193347
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr6_-_133008834
|
0.832
|
NM_138327
|
TAAR1
|
trace amine associated receptor 1
|
|
chr6_-_109795925
|
0.831
|
|
CD164
|
CD164 molecule, sialomucin
|
|
chr6_-_49820030
|
0.819
|
NM_001190986
NM_006061
|
CRISP3
|
cysteine-rich secretory protein 3
|
|
chr17_-_36444626
|
0.819
|
NM_030966
|
KRTAP1-3
|
keratin associated protein 1-3
|
|
chr1_+_246635918
|
0.809
|
NM_030904
|
OR2T1
|
olfactory receptor, family 2, subfamily T, member 1
|
|
chr2_+_33592445
|
0.797
|
NM_015376
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated)
|
|
chr1_-_191422346
|
0.794
|
NM_003783
|
B3GALT2
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2
|
|
chr1_-_103269356
|
0.783
|
|
COL11A1
|
collagen, type XI, alpha 1
|
|
chr16_+_68284252
|
0.780
|
|
NFAT5
|
nuclear factor of activated T-cells 5, tonicity-responsive
|
|
chr15_+_55298908
|
0.776
|
NM_207040
|
TCF12
|
transcription factor 12
|
|
chr6_+_136214495
|
0.772
|
NM_018945
|
PDE7B
|
phosphodiesterase 7B
|
|
chr4_-_20914626
|
0.770
|
NM_147183
|
KCNIP4
|
Kv channel interacting protein 4
|
|
chrX_+_110074124
|
0.768
|
NM_001128166
NM_001128167
|
PAK3
|
p21 protein (Cdc42/Rac)-activated kinase 3
|
|
chr15_-_53996597
|
0.764
|
NM_198400
|
NEDD4
|
neural precursor cell expressed, developmentally down-regulated 4
|
|
chr11_-_16454493
|
0.763
|
NM_033326
|
SOX6
|
SRY (sex determining region Y)-box 6
|
|
chrX_+_14813333
|
0.762
|
NM_001177475
|
MOSPD2
|
motile sperm domain containing 2
|
|
chr15_+_32048380
|
0.754
|
NM_012125
|
CHRM5
|
cholinergic receptor, muscarinic 5
|
|
chr15_-_23201740
|
0.743
|
NM_130838
|
UBE3A
|
ubiquitin protein ligase E3A
|
|
chr8_+_32698892
|
0.740
|
NM_001159996
|
NRG1
|
neuregulin 1
|
|
chr2_+_178893216
|
0.739
|
NM_145739
|
OSBPL6
|
oxysterol binding protein-like 6
|
|
chr5_+_73145098
|
0.729
|
|
RGNEF
|
190 kDa guanine nucleotide exchange factor
|
|
chr21_-_31995972
|
0.721
|
|
SCAF4
|
SR-related CTD-associated factor 4
|
|
chr1_+_202033273
|
0.712
|
NM_001174108
|
ZBED6
|
zinc finger, BED-type containing 6
|
|
chr5_+_71438796
|
0.708
|
NM_005909
|
MAP1B
|
microtubule-associated protein 1B
|
|
chr17_-_36165050
|
0.703
|
NM_181534
|
KRT25
|
keratin 25
|
|
chr15_+_46285789
|
0.697
|
NM_000338
NM_001184832
|
SLC12A1
|
solute carrier family 12 (sodium/potassium/chloride transporters), member 1
|
|
chr7_-_26202626
|
0.695
|
|
HNRNPA2B1
|
heterogeneous nuclear ribonucleoprotein A2/B1
|
|
chrX_+_123062143
|
0.691
|
|
STAG2
|
stromal antigen 2
|
|
chr7_+_113853790
|
0.683
|
NM_001172767
NM_148899
|
FOXP2
|
forkhead box P2
|
|
chr3_-_197726587
|
0.681
|
NM_001077657
|
C3orf43
|
chromosome 3 open reading frame 43
|
|
chr9_+_124836666
|
0.679
|
NM_005294
|
GPR21
|
G protein-coupled receptor 21
|
|
chr6_+_7524691
|
0.678
|
|
DSP
|
desmoplakin
|
|
chr17_-_10265991
|
0.675
|
NM_002472
|
MYH8
|
myosin, heavy chain 8, skeletal muscle, perinatal
|
|
chr12_+_84792203
|
0.673
|
NM_006183
|
NTS
|
neurotensin
|
|
chr12_-_11041740
|
0.664
|
NM_176889
|
TAS2R20
|
taste receptor, type 2, member 20
|
|
chr10_+_77212524
|
0.663
|
NM_032024
|
C10orf11
|
chromosome 10 open reading frame 11
|
|
chrX_+_16578201
|
0.663
|
NM_004057
|
S100G
|
S100 calcium binding protein G
|
|
chr6_+_49575629
|
0.662
|
NM_001010904
|
GLYATL3
|
glycine-N-acyltransferase-like 3
|
|
chr18_-_26996742
|
0.661
|
NM_004948
NM_024421
|
DSC1
|
desmocollin 1
|
|
chr12_-_11178109
|
0.658
|
NM_001097643
|
TAS2R30
|
taste receptor, type 2, member 30
|
|
chrX_-_115508164
|
0.658
|
NM_001017978
|
CXorf61
|
chromosome X open reading frame 61
|
|
chr17_-_36376663
|
0.658
|
NM_213656
|
KRT39
|
keratin 39
|
|
chr18_+_19826734
|
0.653
|
NM_153211
|
TTC39C
|
tetratricopeptide repeat domain 39C
|
|
chr5_-_160211623
|
0.653
|
NM_025153
|
ATP10B
|
ATPase, class V, type 10B
|
|
chr4_-_74307637
|
0.651
|
|
ANKRD17
|
ankyrin repeat domain 17
|
|
chr3_+_35658852
|
0.650
|
NM_016300
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa
|
|
chr12_+_54172413
|
0.650
|
NM_001005519
|
OR6C68
|
olfactory receptor, family 6, subfamily C, member 68
|
|
chr12_+_49155798
|
0.645
|
|
LARP4
|
La ribonucleoprotein domain family, member 4
|
|
chr6_+_101948102
|
0.644
|
|
|
|
|
chr5_+_102493155
|
0.644
|
NM_015216
|
PPIP5K2
|
diphosphoinositol pentakisphosphate kinase 2
|
|
chr22_+_21006814
|
0.638
|
|
|
|
|
chr3_-_56673113
|
0.636
|
NM_015224
|
C3orf63
|
chromosome 3 open reading frame 63
|
|
chr2_-_21081694
|
0.636
|
|
APOB
|
apolipoprotein B (including Ag(x) antigen)
|
|
chr6_+_45404031
|
0.629
|
NM_001015051
NM_001024630
|
RUNX2
|
runt-related transcription factor 2
|
|
chr17_-_61655915
|
0.624
|
NM_000042
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I)
|
|
chr3_-_115960807
|
0.623
|
NM_001164344
NM_001164345
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr9_-_27287106
|
0.622
|
NM_001161585
NM_020641
|
C9orf11
|
chromosome 9 open reading frame 11
|
|
chr6_+_163904564
|
0.617
|
|
QKI
|
quaking homolog, KH domain RNA binding (mouse)
|
|
chr6_-_2696152
|
0.617
|
NM_001012418
|
MYLK4
|
myosin light chain kinase family, member 4
|
|
chr3_-_171786468
|
0.613
|
NM_020949
|
SLC7A14
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 14
|
|
chr13_-_69580459
|
0.608
|
NM_020866
|
KLHL1
|
kelch-like 1 (Drosophila)
|
|
chr16_+_20370359
|
0.605
|
NM_001010845
|
ACSM2A
|
acyl-CoA synthetase medium-chain family member 2A
|
|
chrX_+_55666557
|
0.603
|
NM_198451
|
FOXR2
|
forkhead box R2
|
|
chr12_-_11106159
|
0.602
|
NM_176887
|
TAS2R46
|
taste receptor, type 2, member 46
|
|
chr1_+_165564904
|
0.601
|
NM_001198783
|
POU2F1
|
POU class 2 homeobox 1
|
|
chr14_-_105681377
|
0.596
|
|
IGHV4-31
IGHG1
|
immunoglobulin heavy variable 4-31
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr4_+_46728051
|
0.594
|
NM_000812
|
GABRB1
|
gamma-aminobutyric acid (GABA) A receptor, beta 1
|
|
chr2_+_172252194
|
0.592
|
NM_001378
|
DYNC1I2
|
dynein, cytoplasmic 1, intermediate chain 2
|
|
chr6_+_157141565
|
0.591
|
|
ARID1B
|
AT rich interactive domain 1B (SWI1-like)
|
|
chr12_-_11030777
|
0.588
|
NM_176890
|
TAS2R50
|
taste receptor, type 2, member 50
|
|
chr12_-_78831821
|
0.583
|
NM_001143886
|
PPP1R12A
|
protein phosphatase 1, regulatory (inhibitor) subunit 12A
|
|
chr4_+_70896236
|
0.582
|
NM_001009181
NM_003154
|
STATH
|
statherin
|
|
chr2_+_102456193
|
0.582
|
NM_001011552
|
SLC9A4
|
solute carrier family 9 (sodium/hydrogen exchanger), member 4
|
|
chrX_-_33267646
|
0.581
|
NM_000109
|
DMD
|
dystrophin
|
|
chr13_-_102517196
|
0.580
|
NM_000452
|
SLC10A2
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 2
|
|
chr1_+_157067729
|
0.578
|
NM_002432
|
MNDA
|
myeloid cell nuclear differentiation antigen
|
|
chr11_-_5766621
|
0.575
|
NM_001001913
|
OR52N1
|
olfactory receptor, family 52, subfamily N, member 1
|
|
chr4_-_153523113
|
0.572
|
NM_001013415
|
FBXW7
|
F-box and WD repeat domain containing 7
|
|
chr13_+_32488477
|
0.571
|
NM_004795
|
KL
|
klotho
|
|
chr21_-_26175068
|
0.571
|
|
APP
|
amyloid beta (A4) precursor protein
|
|
chr7_+_28418668
|
0.569
|
NM_182898
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr10_-_46039941
|
0.567
|
NM_001042357
NM_001042389
|
PTPN20B
PTPN20A
|
protein tyrosine phosphatase, non-receptor type 20B
protein tyrosine phosphatase, non-receptor type 20A
|
|
chrX_-_108611940
|
0.567
|
NM_001522
|
GUCY2F
|
guanylate cyclase 2F, retinal
|
|
chr21_-_30642794
|
0.564
|
NM_181624
|
KRTAP23-1
|
keratin associated protein 23-1
|
|
chr15_+_91244422
|
0.564
|
NM_001042572
NM_001271
|
CHD2
|
chromodomain helicase DNA binding protein 2
|
|
chr7_-_76667045
|
0.562
|
NM_006682
|
FGL2
|
fibrinogen-like 2
|
|
chr5_-_63293715
|
0.556
|
|
HTR1A
|
5-hydroxytryptamine (serotonin) receptor 1A
|
|
chr3_-_168674502
|
0.556
|
NM_006217
|
SERPINI2
|
serpin peptidase inhibitor, clade I (pancpin), member 2
|
|
chr1_-_157950898
|
0.553
|
NM_000567
|
CRP
|
C-reactive protein, pentraxin-related
|
|
chr6_+_132901119
|
0.552
|
NM_175057
|
TAAR9
|
trace amine associated receptor 9 (gene/pseudogene)
|
|
chr3_-_25654659
|
0.551
|
|
TOP2B
|
topoisomerase (DNA) II beta 180kDa
|
|
chr1_+_145892190
|
0.548
|
NM_001097616
|
GPR89C
|
G protein-coupled receptor 89C
|
|
chr2_-_29150630
|
0.546
|
NM_001029883
|
C2orf71
|
chromosome 2 open reading frame 71
|
|
chr11_+_89624017
|
0.544
|
|
|
|
|
chr13_-_101852028
|
0.542
|
NM_175929
|
FGF14
|
fibroblast growth factor 14
|
|
chr1_-_239865724
|
0.542
|
NM_001821
|
CHML
|
choroideremia-like (Rab escort protein 2)
|
|
chr4_-_44348414
|
0.541
|
NM_182592
|
YIPF7
|
Yip1 domain family, member 7
|
|
chr13_+_51933228
|
0.540
|
|
CKAP2
|
cytoskeleton associated protein 2
|
|
chr1_+_67404756
|
0.539
|
NM_144701
|
IL23R
|
interleukin 23 receptor
|
|
chrX_-_105169370
|
0.535
|
NM_000354
|
SERPINA7
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 7
|
|
chr22_+_21065202
|
0.535
|
|
|
|
|
chr8_-_7207899
|
0.534
|
NM_001164457
|
ZNF705G
|
zinc finger protein 705G
|
|
chr11_+_59953637
|
0.531
|
NM_023945
|
MS4A5
|
membrane-spanning 4-domains, subfamily A, member 5
|
|
chr6_+_144654565
|
0.531
|
NM_007124
|
UTRN
|
utrophin
|
|
chr4_-_64957595
|
0.530
|
NM_001010874
|
TECRL
|
trans-2,3-enoyl-CoA reductase-like
|
|
chrX_+_51562894
|
0.529
|
NM_001005332
|
MAGED1
|
melanoma antigen family D, 1
|
|
chr22_+_21393107
|
0.528
|
|
IGLJ3
|
immunoglobulin lambda joining 3
|
|
chr9_+_71009746
|
0.526
|
NM_001170415
NM_001170416
|
TJP2
|
tight junction protein 2 (zona occludens 2)
|
|
chr6_+_88111289
|
0.526
|
NM_001010868
|
C6orf163
|
chromosome 6 open reading frame 163
|
|
chr11_+_5558682
|
0.526
|
NM_001005162
|
OR52B6
|
olfactory receptor, family 52, subfamily B, member 6
|
|
chr5_+_147775652
|
0.525
|
|
FBXO38
|
F-box protein 38
|
|
chr14_-_34252695
|
0.524
|
NM_021914
|
CFL2
|
cofilin 2 (muscle)
|
|
chr14_-_24173205
|
0.523
|
NM_004131
|
GZMB
|
granzyme B (granzyme 2, cytotoxic T-lymphocyte-associated serine esterase 1)
|
|
chr20_+_29420081
|
0.522
|
NM_054112
|
DEFB118
|
defensin, beta 118
|
|
chr14_-_19553191
|
0.521
|
NM_001004712
|
OR4K14
|
olfactory receptor, family 4, subfamily K, member 14
|
|
chr1_-_245988330
|
0.521
|
NM_012353
|
OR1C1
|
olfactory receptor, family 1, subfamily C, member 1
|
|
chr5_+_42792676
|
0.519
|
NM_001134848
|
CCDC152
|
coiled-coil domain containing 152
|
|
chr10_+_86121526
|
0.516
|
|
FAM190B
|
family with sequence similarity 190, member B
|
|
chr1_+_61642287
|
0.514
|
|
NFIA
|
nuclear factor I/A
|
|
chr15_+_91244563
|
0.513
|
|
CHD2
|
chromodomain helicase DNA binding protein 2
|
|
chr15_-_23204887
|
0.513
|
NM_000462
|
UBE3A
|
ubiquitin protein ligase E3A
|
|
chr5_+_65408500
|
0.511
|
|
ERBB2IP
|
erbb2 interacting protein
|
|
chr17_+_24394281
|
0.511
|
|
PIPOX
|
pipecolic acid oxidase
|
|
chr2_+_234302511
|
0.510
|
NM_019093
|
UGT1A3
|
UDP glucuronosyltransferase 1 family, polypeptide A3
|
|
chr22_+_21065134
|
0.510
|
|
LOC100290481
|
immunoglobulin lambda light chain-like
|
|
chr1_+_165565006
|
0.510
|
|
POU2F1
|
POU class 2 homeobox 1
|
|
chr4_-_2200755
|
0.508
|
NM_181808
|
POLN
|
polymerase (DNA directed) nu
|
|
chr2_+_86186815
|
0.508
|
NM_017952
|
PTCD3
|
Pentatricopeptide repeat domain 3
|
|
chr4_+_41395493
|
0.507
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr4_-_152368492
|
0.506
|
|
SH3D19
|
SH3 domain containing 19
|
|
chr2_+_171280182
|
0.503
|
|
|
|
|
chr11_+_10729357
|
0.501
|
NM_014633
|
CTR9
|
Ctr9, Paf1/RNA polymerase II complex component, homolog (S. cerevisiae)
|
|
chr8_-_40874485
|
0.501
|
NM_001135731
NM_024645
|
ZMAT4
|
zinc finger, matrin-type 4
|
|
chr4_+_123380569
|
0.495
|
|
KIAA1109
|
KIAA1109
|
|
chr2_+_166034402
|
0.495
|
NM_001172173
|
CSRNP3
|
cysteine-serine-rich nuclear protein 3
|
|
chr2_-_87906400
|
0.494
|
NM_001078170
|
RGPD2
|
RANBP2-like and GRIP domain containing 2
|
|
chr20_-_43601542
|
0.494
|
NM_080827
|
WFDC6
|
WAP four-disulfide core domain 6
|
|
chr18_-_55136673
|
0.493
|
NM_181654
|
CPLX4
|
complexin 4
|
|
chr16_+_24475129
|
0.487
|
|
RBBP6
|
retinoblastoma binding protein 6
|
|
chr6_-_76259976
|
0.487
|
NM_015687
|
FILIP1
|
filamin A interacting protein 1
|
|
chr20_+_5935897
|
0.485
|
NM_001127458
|
CRLS1
|
cardiolipin synthase 1
|
|
chr12_-_9123126
|
0.483
|
|
A2M
|
alpha-2-macroglobulin
|