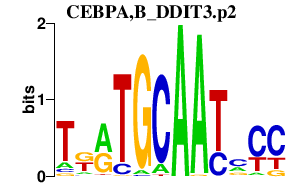
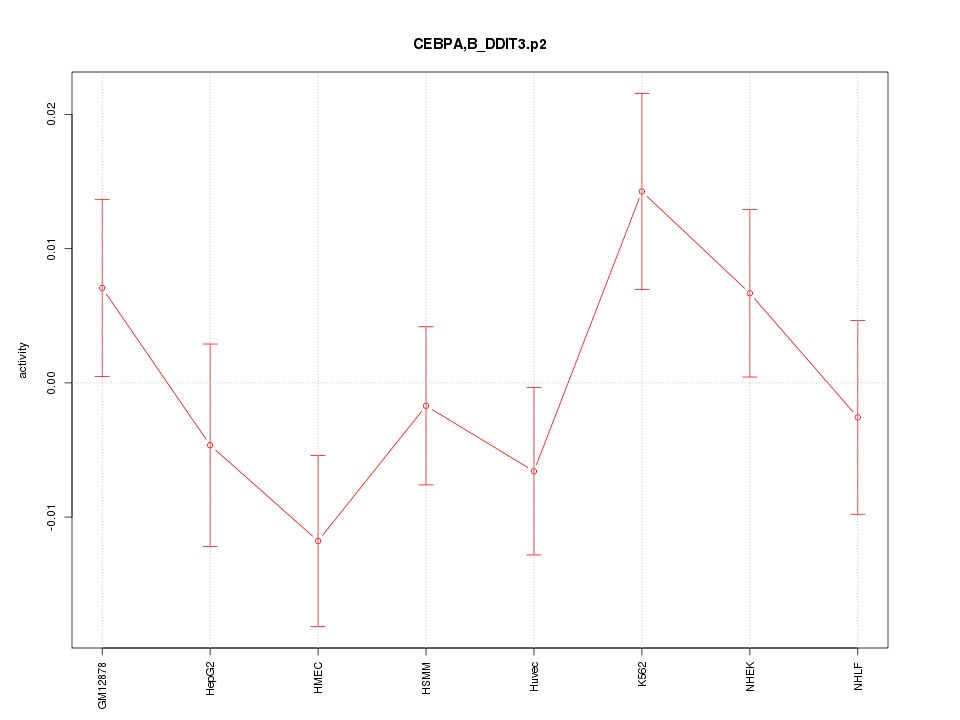
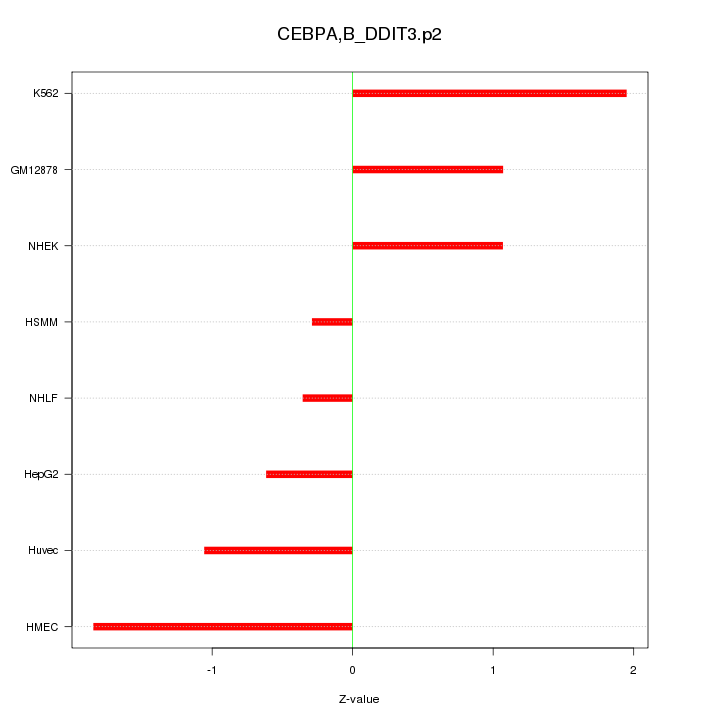
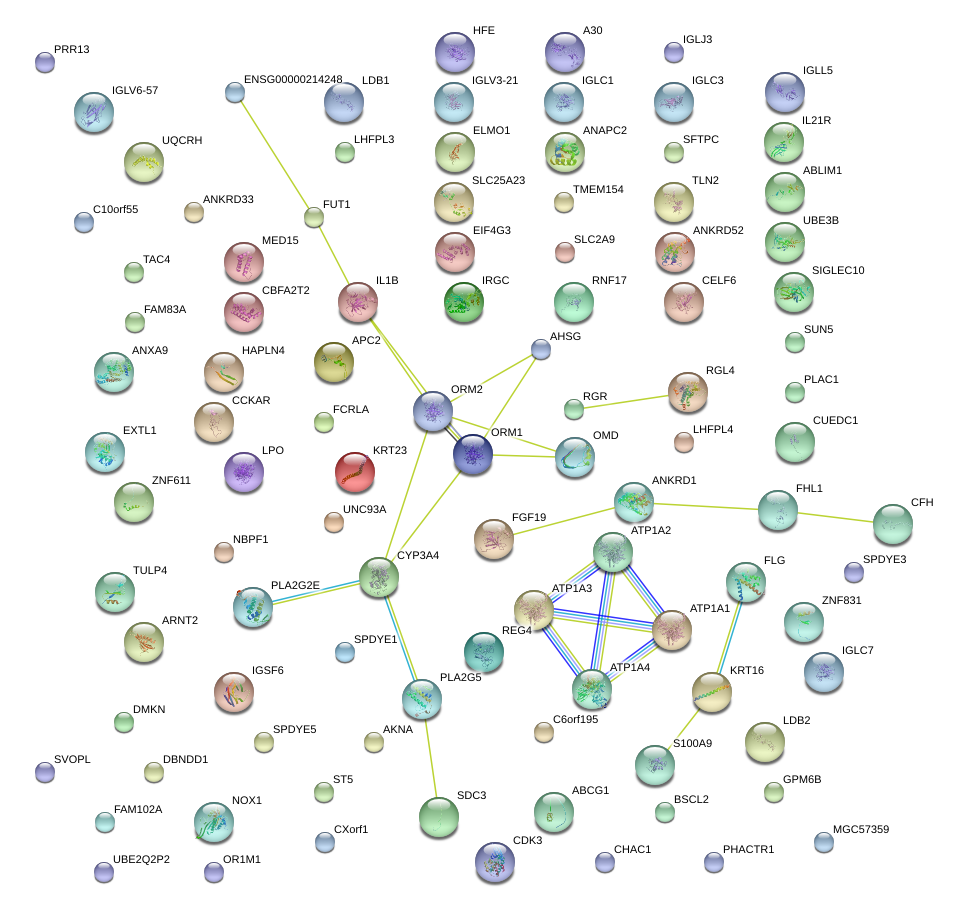
|
chr9_+_116125123
|
2.005
|
NM_000607
|
ORM1
|
orosomucoid 1
|
|
chr17_-_36346240
|
1.768
|
|
KRT23
|
keratin 23 (histone deacetylase inducible)
|
|
chr1_+_149221122
|
1.410
|
NM_003568
|
ANXA9
|
annexin A9
|
|
chr6_+_167624792
|
1.228
|
NM_001143947
NM_018974
|
UNC93A
|
unc-93 homolog A (C. elegans)
|
|
chr19_-_40684638
|
1.064
|
NM_001035516
|
DMKN
|
dermokine
|
|
chr6_+_12825022
|
1.013
|
|
PHACTR1
|
phosphatase and actin regulator 1
|
|
chr1_-_20122638
|
0.943
|
NM_014589
|
PLA2G2E
|
phospholipase A2, group IIE
|
|
chr10_+_85994788
|
0.881
|
NM_001012720
NM_001012722
NM_002921
|
RGR
|
retinal G protein coupled receptor
|
|
chr8_+_124263932
|
0.881
|
NM_032899
NM_207006
|
FAM83A
|
family with sequence similarity 83, member A
|
|
chr4_-_153820535
|
0.798
|
NM_152680
|
TMEM154
|
transmembrane protein 154
|
|
chr1_-_21373816
|
0.794
|
NM_001198803
|
EIF4G3
|
eukaryotic translation initiation factor 4 gamma, 3
|
|
chr11_-_69228286
|
0.769
|
NM_005117
|
FGF19
|
fibroblast growth factor 19
|
|
chr6_+_12824873
|
0.753
|
|
PHACTR1
|
phosphatase and actin regulator 1
|
|
chr22_+_21384862
|
0.731
|
|
IGLV3-21
IGLJ3
IGLC1
|
immunoglobulin lambda variable 3-21
immunoglobulin lambda joining 3
immunoglobulin lambda constant 1 (Mcg marker)
|
|
chr17_-_45280377
|
0.726
|
NM_001077503
NM_001077504
NM_001077505
NM_001077506
NM_170685
|
TAC4
|
tachykinin 4 (hemokinin)
|
|
chr16_+_27346079
|
0.703
|
NM_021798
|
IL21R
|
interleukin 21 receptor
|
|
chr8_+_124264054
|
0.700
|
|
FAM83A
|
family with sequence similarity 83, member A
|
|
chrX_-_133620103
|
0.688
|
NM_021796
|
PLAC1
|
placenta-specific 1
|
|
chr7_+_74962234
|
0.674
|
NM_001099435
|
SPDYE5
|
speedy homolog E5 (Xenopus laevis)
|
|
chr7_-_104354325
|
0.660
|
|
LOC723809
|
hypothetical LOC723809
|
|
chrX_-_24241352
|
0.615
|
NM_001136233
|
FAM48B2
|
family with sequence similarity 48, member B2
|
|
chr8_+_22075112
|
0.610
|
NM_001172357
NM_001172410
NM_003018
|
SFTPC
|
surfactant protein C
|
|
chr12_+_108452117
|
0.583
|
|
UBE3B
|
ubiquitin protein ligase E3B
|
|
chr1_-_31117948
|
0.579
|
|
SDC3
|
syndecan 3
|
|
chr21_+_42492867
|
0.566
|
NM_207627
NM_207628
|
ABCG1
|
ATP-binding cassette, sub-family G (WHITE), member 1
|
|
chr22_+_21079355
|
0.563
|
|
|
|
|
chr4_-_9632192
|
0.558
|
NM_020041
|
SLC2A9
|
solute carrier family 2 (facilitated glucose transporter), member 9
|
|
chr15_-_70385709
|
0.554
|
NM_001172685
|
CELF6
|
CUGBP, Elav-like family member 6
|
|
chr15_+_78520634
|
0.552
|
|
ARNT2
|
aryl-hydrocarbon receptor nuclear translocator 2
|
|
chr9_-_129752687
|
0.538
|
NM_203305
|
FAM102A
|
family with sequence similarity 102, member A
|
|
chrX_-_100015783
|
0.529
|
NM_007052
NM_013955
|
NOX1
|
NADPH oxidase 1
|
|
chr1_+_557269
|
0.520
|
|
|
|
|
chr12_+_52121699
|
0.512
|
NM_001005354
NM_018457
|
PRR13
|
proline rich 13
|
|
chr6_+_158654890
|
0.501
|
|
|
|
|
chr9_-_116196505
|
0.500
|
NM_030767
|
AKNA
|
AT-hook transcription factor
|
|
chr17_-_53335748
|
0.497
|
NM_017949
|
CUEDC1
|
CUE domain containing 1
|
|
chr1_+_26220895
|
0.487
|
|
EXTL1
|
exostoses (multiple)-like 1
|
|
chr19_+_48912053
|
0.486
|
NM_019612
|
IRGC
|
immunity-related GTPase family, cinema
|
|
chr19_+_9064854
|
0.473
|
NM_001004456
|
OR1M1
|
olfactory receptor, family 1, subfamily M, member 1
|
|
chr6_-_2580296
|
0.463
|
NM_152554
|
C6orf195
|
chromosome 6 open reading frame 195
|
|
chr1_+_559129
|
0.447
|
|
|
|
|
chr16_-_21571463
|
0.446
|
NM_005849
|
IGSF6
|
immunoglobulin superfamily, member 6
|
|
chr15_+_80741876
|
0.446
|
|
LOC80154
|
hypothetical LOC80154
|
|
chr20_+_57199469
|
0.441
|
NM_178457
|
ZNF831
|
zinc finger protein 831
|
|
chr15_+_80741827
|
0.439
|
|
LOC80154
|
hypothetical LOC80154
|
|
chr22_+_20880159
|
0.432
|
|
IGLV6-57
|
immunoglobulin lambda variable 6-57
|
|
chr7_+_44007012
|
0.426
|
NM_175064
|
SPDYE1
|
speedy homolog E1 (Xenopus laevis)
|
|
chr17_-_37022465
|
0.425
|
NM_005557
|
KRT16
|
keratin 16
|
|
chr12_+_50568010
|
0.424
|
NM_001130015
NM_182608
|
ANKRD33
|
ankyrin repeat domain 33
|
|
chr22_+_19248810
|
0.417
|
|
MED15
|
mediator complex subunit 15
|
|
chrX_+_135079461
|
0.411
|
NM_001167819
|
FHL1
|
four and a half LIM domains 1
|
|
chr20_-_31055892
|
0.411
|
NM_080675
|
SUN5
|
Sad1 and UNC84 domain containing 5
|
|
chr1_+_158387975
|
0.407
|
NM_144699
|
ATP1A4
|
ATPase, Na+/K+ transporting, alpha 4 polypeptide
|
|
chr10_-_75352540
|
0.405
|
NM_001001791
|
C10orf55
|
chromosome 10 open reading frame 55
|
|
chr11_-_8695974
|
0.403
|
|
ST5
|
suppression of tumorigenicity 5
|
|
chr19_-_56612653
|
0.394
|
NM_001171156
NM_001171157
NM_001171158
NM_001171159
NM_001171160
NM_001171161
NM_033130
|
SIGLEC10
|
sialic acid binding Ig-like lectin 10
|
|
chr1_+_151596953
|
0.386
|
NM_002965
|
S100A9
|
S100 calcium binding protein A9
|
|
chr11_-_62218759
|
0.381
|
|
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin)
|
|
chr6_+_158654464
|
0.380
|
|
TULP4
|
tubby like protein 4
|
|
chr3_-_9570412
|
0.371
|
NM_198560
|
LHFPL4
|
lipoma HMGIC fusion partner-like 4
|
|
chr1_+_116744992
|
0.366
|
|
ATP1A1
|
ATPase, Na+/K+ transporting, alpha 1 polypeptide
|
|
chr20_+_31613800
|
0.350
|
NM_001039709
NM_005093
|
CBFA2T2
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 2
|
|
chr19_-_46504831
|
0.349
|
|
|
|
|
chr16_-_88604029
|
0.346
|
NM_024043
|
DBNDD1
|
dysbindin (dystrobrevin binding protein 1) domain containing 1
|
|
chr16_-_30694799
|
0.341
|
|
|
|
|
chr22_+_20880112
|
0.341
|
|
|
|
|
chr15_+_39032927
|
0.340
|
NM_001142776
NM_024111
|
CHAC1
|
ChaC, cation transport regulator homolog 1 (E. coli)
|
|
chr9_-_94226380
|
0.340
|
NM_005014
|
OMD
|
osteomodulin
|
|
chr15_-_70838703
|
0.337
|
|
|
|
|
chr2_-_113310796
|
0.334
|
NM_000576
|
IL1B
|
interleukin 1, beta
|
|
chr19_+_1401117
|
0.334
|
NM_005883
|
APC2
|
adenomatosis polyposis coli 2
|
|
chr7_-_138014329
|
0.330
|
NM_001139456
|
SVOPL
|
SVOP-like
|
|
chr19_+_1401208
|
0.328
|
|
APC2
|
adenomatosis polyposis coli 2
|
|
chr6_+_37511383
|
0.327
|
|
|
|
|
chr19_-_19234559
|
0.327
|
NM_023002
|
HAPLN4
|
hyaluronan and proteoglycan link protein 4
|
|
chr17_+_53670785
|
0.324
|
NM_001160102
NM_006151
|
LPO
|
lactoperoxidase
|
|
chr7_-_36991189
|
0.322
|
NM_001039459
|
ELMO1
|
engulfment and cell motility 1
|
|
chrX_-_13745108
|
0.318
|
|
GPM6B
|
glycoprotein M6B
|
|
chr1_-_16006779
|
0.315
|
NM_001089591
|
UQCRHL
|
ubiquinol-cytochrome c reductase hinge protein-like
|
|
chr10_-_116434364
|
0.308
|
NM_001003407
NM_001003408
|
ABLIM1
|
actin binding LIM protein 1
|
|
chr19_-_57924946
|
0.308
|
NM_030972
|
ZNF611
|
zinc finger protein 611
|
|
chr22_+_21393107
|
0.305
|
|
IGLJ3
|
immunoglobulin lambda joining 3
|
|
chr6_+_26195426
|
0.305
|
NM_000410
NM_139003
NM_139004
NM_139006
NM_139007
NM_139008
NM_139009
NM_139010
NM_139011
|
HFE
|
hemochromatosis
|
|
chr7_+_99743260
|
0.304
|
NM_001004351
|
SPDYE3
|
speedy homolog E3 (Xenopus laevis)
|
|
chrX_+_144716619
|
0.299
|
NM_004709
|
CXorf1
|
chromosome X open reading frame 1
|
|
chr4_-_26100986
|
0.295
|
NM_000730
|
CCKAR
|
cholecystokinin A receptor
|
|
chr1_+_159943385
|
0.295
|
NM_001184866
NM_001184867
NM_001184870
NM_001184871
NM_001184872
NM_001184873
NM_032738
|
FCRLA
|
Fc receptor-like A
|
|
chr1_+_20269239
|
0.290
|
NM_000929
|
PLA2G5
|
phospholipase A2, group V
|
|
chr15_+_80434362
|
0.282
|
|
UBE2Q2P2
|
ubiquitin-conjugating enzyme E2Q family member 2 pseudogene 2
|
|
chr10_-_103864651
|
0.270
|
NM_003893
|
LDB1
|
LIM domain binding 1
|
|
chr3_+_187813553
|
0.265
|
|
AHSG
|
alpha-2-HS-glycoprotein
|
|
chr10_-_92671005
|
0.263
|
NM_014391
|
ANKRD1
|
ankyrin repeat domain 1 (cardiac muscle)
|
|
chr1_-_120155581
|
0.252
|
NM_001159352
NM_001159353
NM_032044
|
REG4
|
regenerating islet-derived family, member 4
|
|
chr17_+_71508581
|
0.247
|
NM_001258
|
CDK3
|
cyclin-dependent kinase 3
|
|
chr19_-_53950458
|
0.247
|
NM_000148
|
FUT1
|
fucosyltransferase 1 (galactoside 2-alpha-L-fucosyltransferase, H blood group)
|
|
chrX_-_13745134
|
0.246
|
|
GPM6B
|
glycoprotein M6B
|
|
chr15_+_60838053
|
0.245
|
|
TLN2
|
talin 2
|
|
chr10_-_73167586
|
0.243
|
NM_001168390
|
C10orf105
|
chromosome 10 open reading frame 105
|
|
chr22_+_19248733
|
0.243
|
|
MED15
|
mediator complex subunit 15
|
|
chr9_+_135994734
|
0.237
|
NM_052821
|
WDR5
|
WD repeat domain 5
|
|
chr7_-_104696678
|
0.235
|
NM_182691
|
SRPK2
|
SRSF protein kinase 2
|
|
chr6_-_33862690
|
0.233
|
NM_001143944
|
LEMD2
|
LEM domain containing 2
|
|
chr12_-_120781002
|
0.231
|
NM_002150
|
HPD
|
4-hydroxyphenylpyruvate dioxygenase
|
|
chr17_+_4283967
|
0.231
|
NM_182538
|
SPNS3
|
spinster homolog 3 (Drosophila)
|
|
chr17_+_27617307
|
0.227
|
NM_138328
|
RHBDL3
|
rhomboid, veinlet-like 3 (Drosophila)
|
|
chr6_+_32713160
|
0.225
|
NM_002122
|
HLA-DQA1
|
major histocompatibility complex, class II, DQ alpha 1
|
|
chr1_+_111571803
|
0.224
|
NM_001025197
NM_004000
|
CHI3L2
|
chitinase 3-like 2
|
|
chr10_+_69539255
|
0.224
|
NM_032578
|
MYPN
|
myopalladin
|
|
chr10_-_116434102
|
0.220
|
|
ABLIM1
|
actin binding LIM protein 1
|
|
chr11_-_16953135
|
0.219
|
|
RPL36A
|
ribosomal protein L36a
|
|
chr1_-_51660328
|
0.219
|
NM_001159969
|
EPS15
|
epidermal growth factor receptor pathway substrate 15
|
|
chr3_+_141879555
|
0.218
|
NM_152616
|
TRIM42
|
tripartite motif containing 42
|
|
chr18_-_53440005
|
0.212
|
|
NARS
|
asparaginyl-tRNA synthetase
|
|
chr6_+_135558572
|
0.211
|
|
|
|
|
chr1_-_116185215
|
0.210
|
NM_005599
|
NHLH2
|
nescient helix loop helix 2
|
|
chr13_+_28496746
|
0.210
|
NM_001033602
|
MTUS2
|
microtubule associated tumor suppressor candidate 2
|
|
chr17_+_29707583
|
0.209
|
NM_005408
|
CCL13
|
chemokine (C-C motif) ligand 13
|
|
chr10_-_79459296
|
0.209
|
NM_007055
|
POLR3A
|
polymerase (RNA) III (DNA directed) polypeptide A, 155kDa
|
|
chr17_+_75766875
|
0.206
|
NM_024110
|
CARD14
|
caspase recruitment domain family, member 14
|
|
chr12_-_55404530
|
0.205
|
|
NACA
|
nascent polypeptide-associated complex alpha subunit
|
|
chr15_+_40481879
|
0.205
|
NM_173088
|
CAPN3
|
calpain 3, (p94)
|
|
chr5_+_149131695
|
0.205
|
NM_001172699
|
PPARGC1B
|
peroxisome proliferator-activated receptor gamma, coactivator 1 beta
|
|
chr8_-_27171852
|
0.204
|
|
STMN4
|
stathmin-like 4
|
|
chr8_-_110730183
|
0.200
|
NM_001099747
NM_001099748
NM_001099749
NM_001099750
NM_017786
|
SYBU
|
syntabulin (syntaxin-interacting)
|
|
chr2_+_113542017
|
0.197
|
NM_173161
|
IL1F10
|
interleukin 1 family, member 10 (theta)
|
|
chr15_-_61913134
|
0.195
|
|
HERC1
|
hect (homologous to the E6-AP (UBE3A) carboxyl terminus) domain and RCC1 (CHC1)-like domain (RLD) 1
|
|
chr10_-_92670764
|
0.194
|
|
ANKRD1
|
ankyrin repeat domain 1 (cardiac muscle)
|
|
chr15_+_65217412
|
0.194
|
NM_001145103
|
SMAD3
|
SMAD family member 3
|
|
chr20_-_35318690
|
0.193
|
NM_001184731
NM_021081
|
GHRH
|
growth hormone releasing hormone
|
|
chr9_+_134927185
|
0.192
|
NM_001807
|
CEL
|
carboxyl ester lipase (bile salt-stimulated lipase)
|
|
chr8_-_27171814
|
0.191
|
NM_030795
|
STMN4
|
stathmin-like 4
|
|
chr3_-_150533948
|
0.190
|
NM_001184723
NM_138786
|
TM4SF18
|
transmembrane 4 L six family member 18
|
|
chr16_+_24174374
|
0.190
|
NM_006539
|
CACNG3
|
calcium channel, voltage-dependent, gamma subunit 3
|
|
chr19_-_59368625
|
0.190
|
NM_001145303
NM_144686
|
TMC4
|
transmembrane channel-like 4
|
|
chr18_-_53440012
|
0.189
|
|
NARS
|
asparaginyl-tRNA synthetase
|
|
chr17_-_61655915
|
0.187
|
NM_000042
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I)
|
|
chr19_-_60272708
|
0.187
|
NM_138412
|
RDH13
|
retinol dehydrogenase 13 (all-trans/9-cis)
|
|
chr1_+_17779528
|
0.186
|
NM_001011722
|
ARHGEF10L
|
Rho guanine nucleotide exchange factor (GEF) 10-like
|
|
chr3_-_49701085
|
0.185
|
NM_020998
|
MST1
|
macrophage stimulating 1 (hepatocyte growth factor-like)
|
|
chr17_+_18588050
|
0.185
|
NM_031456
|
FBXW10
CDRT1
|
F-box and WD repeat domain containing 10
CMT1A duplicated region transcript 1
|
|
chr8_+_42671718
|
0.184
|
NM_000749
|
CHRNB3
|
cholinergic receptor, nicotinic, beta 3
|
|
chr11_+_63060848
|
0.184
|
NM_004585
|
RARRES3
|
retinoic acid receptor responder (tazarotene induced) 3
|
|
chr6_-_157664464
|
0.184
|
|
C6orf35
|
chromosome 6 open reading frame 35
|
|
chr1_-_159543847
|
0.182
|
|
MPZ
|
myelin protein zero
|
|
chr16_-_28457825
|
0.182
|
NM_001042483
NM_012385
|
NUPR1
|
nuclear protein, transcriptional regulator, 1
|
|
chr6_+_32003180
|
0.180
|
NM_000063
NM_001145903
|
C2
|
complement component 2
|
|
chr16_-_21571409
|
0.180
|
|
IGSF6
|
immunoglobulin superfamily, member 6
|
|
chr6_+_135558744
|
0.180
|
|
MYB
|
v-myb myeloblastosis viral oncogene homolog (avian)
|
|
chr11_+_18244383
|
0.180
|
NM_000331
NM_001178006
NM_199161
|
SAA1
|
serum amyloid A1
|
|
chr11_-_10487212
|
0.180
|
NM_001190702
|
MTRNR2L8
|
MT-RNR2-like 8
|
|
chr1_-_180822686
|
0.179
|
NM_021133
|
RNASEL
|
ribonuclease L (2',5'-oligoisoadenylate synthetase-dependent)
|
|
chr17_-_75427644
|
0.177
|
NM_003655
|
CBX4
|
chromobox homolog 4
|
|
chr17_-_77488258
|
0.174
|
NM_006907
NM_153824
|
PYCR1
|
pyrroline-5-carboxylate reductase 1
|
|
chrX_+_69314060
|
0.174
|
NM_198512
|
DGAT2L6
|
diacylglycerol O-acyltransferase 2-like 6
|
|
chr7_+_28305464
|
0.171
|
NM_182899
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr2_+_169465995
|
0.169
|
NM_001081686
NM_021176
|
G6PC2
|
glucose-6-phosphatase, catalytic, 2
|
|
chr4_+_39874921
|
0.168
|
NM_004310
|
RHOH
|
ras homolog gene family, member H
|
|
chr9_-_76692829
|
0.168
|
NM_017662
|
TRPM6
|
transient receptor potential cation channel, subfamily M, member 6
|
|
chr12_+_121218218
|
0.167
|
NM_152759
|
LRRC43
|
leucine rich repeat containing 43
|
|
chr1_+_26376611
|
0.165
|
|
CNKSR1
|
connector enhancer of kinase suppressor of Ras 1
|
|
chr19_+_60838170
|
0.165
|
|
|
|
|
chrX_+_47871546
|
0.163
|
NM_205856
|
SPACA5
|
sperm acrosome associated 5
|
|
chr9_+_94766379
|
0.157
|
|
FGD3
|
FYVE, RhoGEF and PH domain containing 3
|
|
chr17_-_15463742
|
0.156
|
NM_006382
|
CDRT1
|
CMT1A duplicated region transcript 1
|
|
chr21_-_46428788
|
0.156
|
NM_001142854
NM_032261
|
C21orf56
|
chromosome 21 open reading frame 56
|
|
chr11_+_4571844
|
0.155
|
NM_001005169
|
OR52I1
|
olfactory receptor, family 52, subfamily I, member 1
|
|
chr20_+_62266270
|
0.155
|
NM_004535
|
MYT1
|
myelin transcription factor 1
|
|
chr20_+_54420574
|
0.154
|
NM_001164116
NM_001164114
NM_001164115
NM_020356
|
CASS4
|
Cas scaffolding protein family member 4
|
|
chr10_-_29963847
|
0.153
|
|
|
|
|
chr8_-_38127489
|
0.153
|
NM_000349
|
STAR
|
steroidogenic acute regulatory protein
|
|
chr21_+_29322100
|
0.150
|
|
USP16
|
ubiquitin specific peptidase 16
|
|
chr11_-_65304397
|
0.148
|
NM_138368
|
DKFZp761E198
|
DKFZp761E198 protein
|
|
chr19_+_55614006
|
0.147
|
NM_003121
|
SPIB
|
Spi-B transcription factor (Spi-1/PU.1 related)
|
|
chr1_-_205210567
|
0.146
|
NM_001122979
NM_001170631
NM_032029
|
FCAMR
|
Fc receptor, IgA, IgM, high affinity
|
|
chr12_-_131725848
|
0.146
|
|
POLE
|
polymerase (DNA directed), epsilon
|
|
chr18_-_53439949
|
0.146
|
|
NARS
|
asparaginyl-tRNA synthetase
|
|
chr11_-_118693021
|
0.146
|
NM_006500
|
MCAM
|
melanoma cell adhesion molecule
|
|
chr3_-_42818221
|
0.143
|
|
|
|
|
chr15_-_61913186
|
0.142
|
NM_003922
|
HERC1
|
hect (homologous to the E6-AP (UBE3A) carboxyl terminus) domain and RCC1 (CHC1)-like domain (RLD) 1
|
|
chr3_-_150422474
|
0.142
|
NM_000096
|
CP
|
ceruloplasmin (ferroxidase)
|
|
chr7_-_22828922
|
0.141
|
NM_019059
|
TOMM7
|
translocase of outer mitochondrial membrane 7 homolog (yeast)
|
|
chr1_+_40612891
|
0.141
|
NM_001198978
|
SMAP2
|
small ArfGAP2
|
|
chrX_-_102828238
|
0.140
|
NM_001142418
NM_001142423
NM_001142424
NM_001142425
NM_001142426
NM_001142429
NM_001142430
NM_012286
|
MORF4L2
|
mortality factor 4 like 2
|
|
chr2_+_237143118
|
0.139
|
NM_020311
|
CXCR7
|
chemokine (C-X-C motif) receptor 7
|
|
chr7_+_129720209
|
0.139
|
NM_001163446
NM_016352
|
CPA4
|
carboxypeptidase A4
|
|
chr17_+_3326045
|
0.139
|
NM_000049
|
ASPA
|
aspartoacylase
|
|
chrX_+_55043447
|
0.136
|
NM_014481
|
APEX2
|
APEX nuclease (apurinic/apyrimidinic endonuclease) 2
|
|
chr1_-_9040141
|
0.134
|
|
SLC2A5
|
solute carrier family 2 (facilitated glucose/fructose transporter), member 5
|
|
chr19_+_13111072
|
0.133
|
|
NACC1
|
nucleus accumbens associated 1, BEN and BTB (POZ) domain containing
|
|
chr6_-_39180390
|
0.133
|
|
C6orf64
|
chromosome 6 open reading frame 64
|
|
chr3_+_187813405
|
0.133
|
|
AHSG
|
alpha-2-HS-glycoprotein
|
|
chr15_-_64108718
|
0.133
|
|
|
|
|
chr19_-_61876057
|
0.132
|
NM_001005850
|
ZNF835
|
zinc finger protein 835
|
|
chrX_-_15198509
|
0.132
|
NM_001168531
|
ASB9
|
ankyrin repeat and SOCS box containing 9
|
|
chr1_+_203106600
|
0.131
|
|
NFASC
|
neurofascin
|
|
chr19_-_51889002
|
0.131
|
|
PRKD2
|
protein kinase D2
|
|
chr1_+_234748187
|
0.131
|
NM_201545
|
LGALS8
|
lectin, galactoside-binding, soluble, 8
|
|
chr1_-_151549790
|
0.130
|
NM_052891
|
PGLYRP3
|
peptidoglycan recognition protein 3
|
|
chr1_-_6448737
|
0.129
|
NM_001039664
NM_003790
NM_148965
NM_148966
NM_148967
NM_148970
|
TNFRSF25
|
tumor necrosis factor receptor superfamily, member 25
|
|
chrX_-_13744933
|
0.129
|
|
GPM6B
|
glycoprotein M6B
|