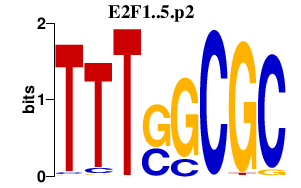
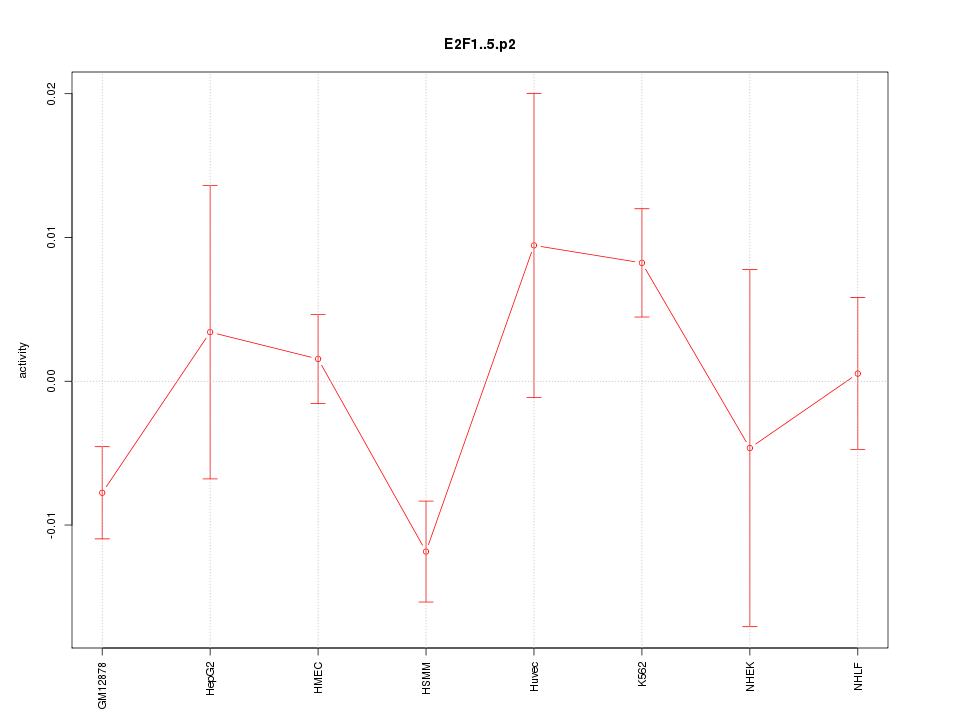
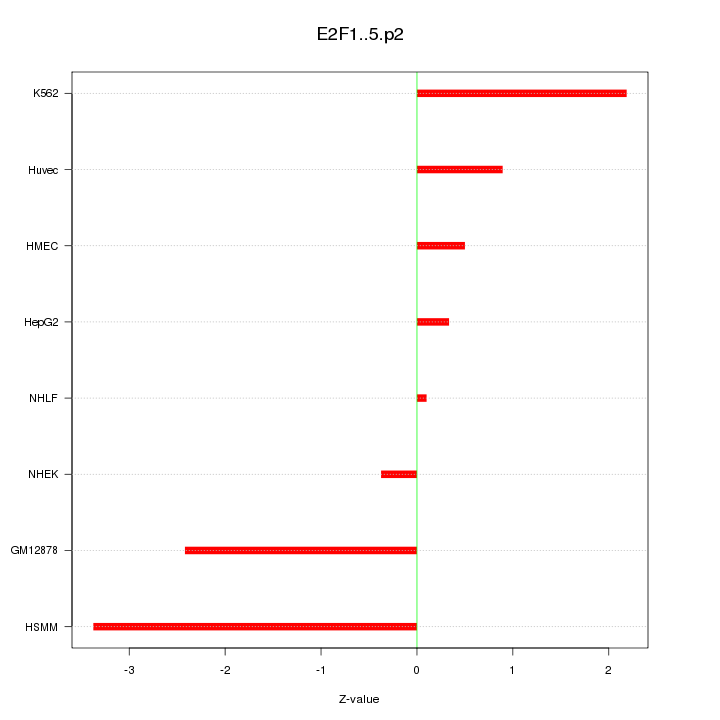
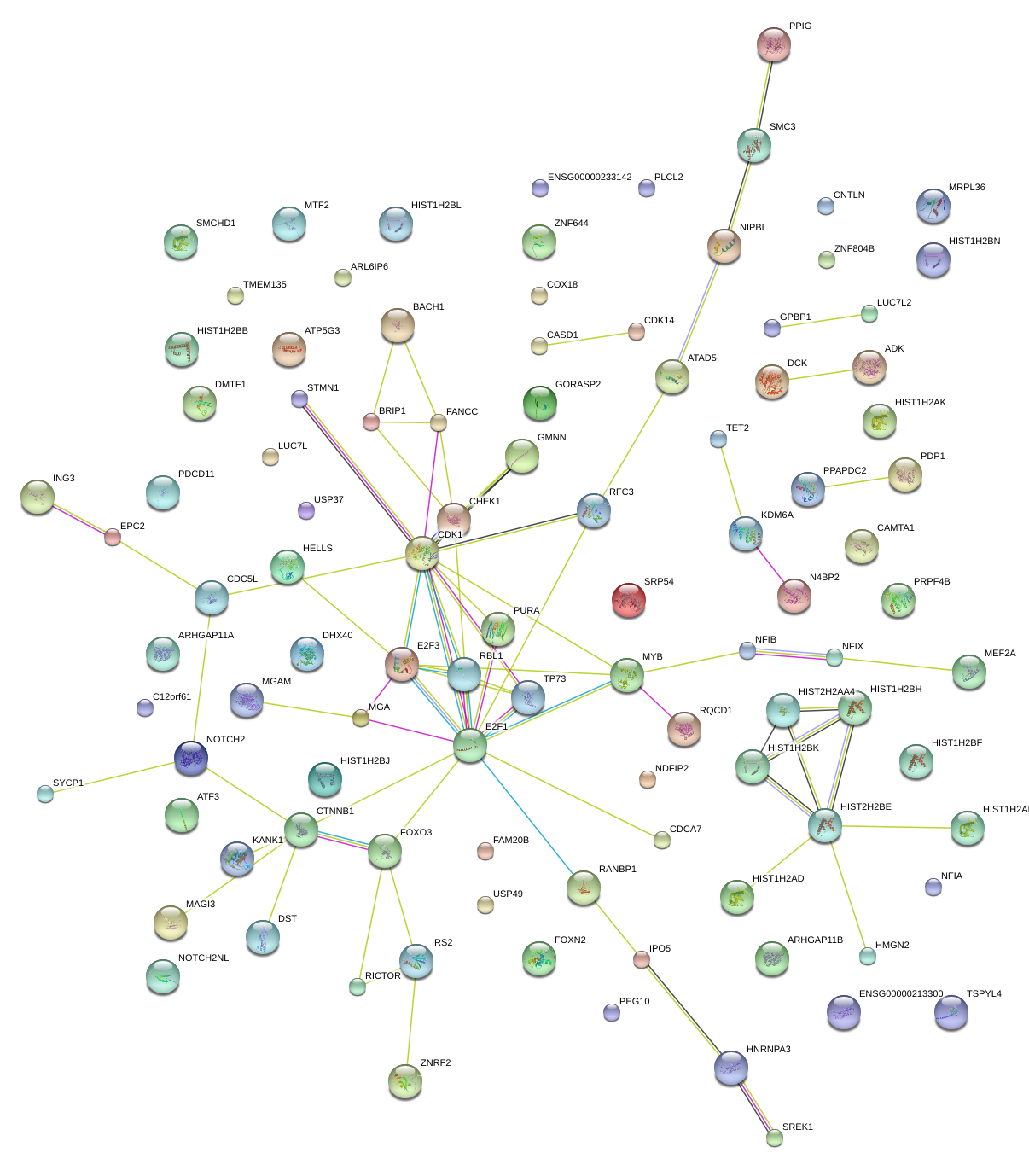
|
chr7_+_120378052
|
2.915
|
NM_019071
NM_198267
|
ING3
|
inhibitor of growth family, member 3
|
|
chr10_+_62208256
|
2.545
|
|
CDK1
|
cyclin-dependent kinase 1
|
|
chr10_+_62208094
|
2.536
|
NM_001170406
NM_001170407
NM_001786
NM_033379
|
CDK1
|
cyclin-dependent kinase 1
|
|
chr2_+_173927783
|
2.156
|
NM_031942
NM_145810
|
CDCA7
|
cell division cycle associated 7
|
|
chr10_+_96295511
|
1.952
|
NM_018063
|
HELLS
|
helicase, lymphoid-specific
|
|
chr1_+_143920424
|
1.787
|
NM_203458
|
NOTCH2NL
|
notch 2 N-terminal like
|
|
chr7_+_93977267
|
1.605
|
|
CASD1
|
CAS1 domain containing 1
|
|
chr6_+_20510019
|
1.595
|
NM_001949
|
E2F3
|
E2F transcription factor 3
|
|
chr11_+_86426518
|
1.594
|
NM_001168724
NM_022918
|
TMEM135
|
transmembrane protein 135
|
|
chr1_+_26671548
|
1.552
|
|
HMGN2
|
high-mobility group nucleosomal binding domain 2
|
|
chr5_+_139473873
|
1.482
|
NM_005859
|
PURA
|
purine-rich element binding protein A
|
|
chrX_+_44617346
|
1.476
|
NM_021140
|
KDM6A
|
lysine (K)-specific demethylase 6A
|
|
chr2_+_48395271
|
1.464
|
NM_002158
|
FOXN2
|
forkhead box N2
|
|
chr1_+_113734997
|
1.423
|
NM_001142782
NM_152900
|
MAGI3
|
membrane associated guanylate kinase, WW and PDZ domain containing 3
|
|
chr5_+_56505608
|
1.419
|
|
GPBP1
|
GC-rich promoter binding protein 1
|
|
chr1_+_210848726
|
1.395
|
|
ATF3
|
activating transcription factor 3
|
|
chr1_+_210848763
|
1.345
|
|
ATF3
|
activating transcription factor 3
|
|
chr15_+_97923655
|
1.303
|
NM_001130926
NM_001130927
NM_001171894
NM_005587
|
MEF2A
|
myocyte enhancer factor 2A
|
|
chr13_+_97426713
|
1.293
|
|
IPO5
|
importin 5
|
|
chr1_+_6767968
|
1.260
|
NM_001195563
NM_015215
|
CAMTA1
|
calmodulin binding transcription activator 1
|
|
chr7_+_93976927
|
1.250
|
NM_022900
|
CASD1
|
CAS1 domain containing 1
|
|
chr13_+_33290254
|
1.246
|
|
RFC3
|
replication factor C (activator 1) 3, 38kDa
|
|
chr6_+_108988797
|
1.238
|
|
FOXO3
|
forkhead box O3
|
|
chr7_+_138695144
|
1.233
|
NM_016019
|
LUC7L2
|
LUC7-like 2 (S. cerevisiae)
|
|
chr15_+_97923702
|
1.226
|
|
MEF2A
|
myocyte enhancer factor 2A
|
|
chr10_+_112317401
|
1.209
|
NM_005445
|
SMC3
|
structural maintenance of chromosomes 3
|
|
chr1_+_93317374
|
1.198
|
NM_001164391
NM_001164392
NM_001164393
NM_007358
|
MTF2
|
metal response element binding transcription factor 2
|
|
chr7_+_32734244
|
1.194
|
|
LOC441208
|
zinc and ring finger 2 pseudogene
|
|
chr6_+_3966499
|
1.166
|
NM_003913
|
PRPF4B
|
PRP4 pre-mRNA processing factor 4 homolog B (yeast)
|
|
chr1_+_3558958
|
1.164
|
NM_005427
|
TP73
|
tumor protein p73
|
|
chr3_+_16901455
|
1.164
|
NM_001144382
|
PLCL2
|
phospholipase C-like 2
|
|
chr9_-_97119677
|
1.156
|
NM_000136
|
FANCC
|
Fanconi anemia, complementation group C
|
|
chr7_+_138695209
|
1.152
|
|
LUC7L2
|
LUC7-like 2 (S. cerevisiae)
|
|
chr6_+_24883133
|
1.143
|
NM_015895
|
GMNN
|
geminin, DNA replication inhibitor
|
|
chr15_+_97923737
|
1.129
|
|
MEF2A
|
myocyte enhancer factor 2A
|
|
chr9_+_17124974
|
1.115
|
NM_001114395
NM_017738
|
CNTLN
|
centlein, centrosomal protein
|
|
chr5_-_39110235
|
1.108
|
|
RICTOR
|
RPTOR independent companion of MTOR, complex 2
|
|
chr17_+_26183074
|
1.108
|
NM_024857
|
ATAD5
|
ATPase family, AAA domain containing 5
|
|
chr1_+_210848592
|
1.107
|
NM_001040619
NM_001674
|
ATF3
|
activating transcription factor 3
|
|
chr2_+_219141761
|
1.103
|
NM_005444
|
RQCD1
|
RCD1 required for cell differentiation1 homolog (S. pombe)
|
|
chr4_+_106287391
|
1.096
|
NM_001127208
NM_017628
|
TET2
|
tet oncogene family member 2
|
|
chr3_+_41215926
|
1.096
|
NM_001098209
NM_001098210
NM_001904
|
CTNNB1
|
catenin (cadherin-associated protein), beta 1, 88kDa
|
|
chr7_+_88226617
|
1.090
|
NM_181646
|
ZNF804B
|
zinc finger protein 804B
|
|
chr5_+_36912656
|
1.090
|
|
NIPBL
|
Nipped-B homolog (Drosophila)
|
|
chr7_+_138695236
|
1.082
|
|
LUC7L2
|
LUC7-like 2 (S. cerevisiae)
|
|
chr20_-_35157744
|
1.079
|
|
RBL1
|
retinoblastoma-like 1 (p107)
|
|
chr12_-_61283474
|
1.066
|
NM_175895
|
C12orf61
|
chromosome 12 open reading frame 61
|
|
chr17_+_54997667
|
1.051
|
NM_001166301
NM_024612
|
DHX40
|
DEAH (Asp-Glu-Ala-His) box polypeptide 40
|
|
chr9_+_494661
|
1.042
|
NM_015158
|
KANK1
|
KN motif and ankyrin repeat domains 1
|
|
chr5_-_39110251
|
1.027
|
NM_152756
|
RICTOR
|
RPTOR independent companion of MTOR, complex 2
|
|
chr5_+_65476418
|
1.016
|
NM_139168
|
SREK1
|
splicing regulatory glutamine/lysine-rich protein 1
|
|
chr7_+_138695241
|
1.000
|
|
LUC7L2
|
LUC7-like 2 (S. cerevisiae)
|
|
chr2_-_175754366
|
0.978
|
NM_001002258
NM_001190329
NM_001689
|
ATP5G3
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C3 (subunit 9)
|
|
chr4_+_72078126
|
0.972
|
NM_000788
|
DCK
|
deoxycytidine kinase
|
|
chr6_-_27914095
|
0.972
|
NM_003510
|
HIST1H2AK
|
histone cluster 1, H2ak
|
|
chr2_+_131966970
|
0.970
|
|
LOC150776
|
sphingomyelin phosphodiesterase 4, neutral membrane pseudogene
|
|
chr7_+_64136078
|
0.964
|
|
CCT6P3
|
chaperonin containing TCP1, subunit 6 (zeta) pseudogene 3
|
|
chr1_-_26105230
|
0.961
|
NM_203401
|
STMN1
|
stathmin 1
|
|
chr6_+_27208763
|
0.961
|
NM_021064
|
HIST1H2AG
|
histone cluster 1, H2ag
|
|
chr6_-_41971064
|
0.946
|
NM_018561
|
USP49
|
ubiquitin specific peptidase 49
|
|
chr4_+_72078255
|
0.935
|
|
DCK
|
deoxycytidine kinase
|
|
chr6_-_56816403
|
0.929
|
|
DST
|
dystonin
|
|
chr20_-_35157815
|
0.926
|
NM_002895
NM_183404
|
RBL1
|
retinoblastoma-like 1 (p107)
|
|
chr2_+_170149348
|
0.924
|
|
PPIG
|
peptidylprolyl isomerase G (cyclophilin G)
|
|
chr1_+_115198977
|
0.915
|
NM_003176
|
SYCP1
|
synaptonemal complex protein 1
|
|
chr1_+_169721260
|
0.914
|
NM_015172
|
PRRC2C
|
proline-rich coiled-coil 2C
|
|
chr1_-_91260357
|
0.911
|
|
ZNF644
|
zinc finger protein 644
|
|
chr2_+_177785667
|
0.909
|
NM_194247
|
HNRNPA3P1
HNRNPA3
|
heterogeneous nuclear ribonucleoprotein A3 pseudogene 1
heterogeneous nuclear ribonucleoprotein A3
|
|
chr5_+_56505412
|
0.906
|
NM_022913
|
GPBP1
|
GC-rich promoter binding protein 1
|
|
chr1_+_177261678
|
0.898
|
NM_014864
|
FAM20B
|
family with sequence similarity 20, member B
|
|
chr5_+_78568094
|
0.890
|
|
|
|
|
chr2_+_149118791
|
0.887
|
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila)
|
|
chr6_+_44463418
|
0.884
|
|
CDC5L
|
CDC5 cell division cycle 5-like (S. pombe)
|
|
chr13_+_33290205
|
0.881
|
NM_002915
NM_181558
|
RFC3
|
replication factor C (activator 1) 3, 38kDa
|
|
chr7_+_86619805
|
0.876
|
NM_021145
|
DMTF1
|
cyclin D binding myb-like transcription factor 1
|
|
chr20_-_31737731
|
0.874
|
|
E2F1
|
E2F transcription factor 1
|
|
chr7_+_94123672
|
0.872
|
|
PEG10
|
paternally expressed 10
|
|
chr7_+_86619580
|
0.869
|
NM_001142326
NM_001142327
|
DMTF1
|
cyclin D binding myb-like transcription factor 1
|
|
chr2_+_149119029
|
0.863
|
NM_015630
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila)
|
|
chr14_+_85069217
|
0.856
|
|
|
|
|
chr2_+_171493870
|
0.841
|
NM_015530
|
GORASP2
|
golgi reassembly stacking protein 2, 55kDa
|
|
chr15_+_28705656
|
0.841
|
|
ARHGAP11B
|
Rho GTPase activating protein 11B
|
|
chr8_+_94998258
|
0.841
|
NM_018444
NM_001161780
NM_001161779
NM_001161781
|
PDP1
|
pyruvate dehyrogenase phosphatase catalytic subunit 1
|
|
chr6_-_27208507
|
0.839
|
NM_021058
|
HIST1H2BJ
|
histone cluster 1, H2bj
|
|
chr10_+_75580914
|
0.835
|
NM_006721
|
ADK
|
adenosine kinase
|
|
chr1_-_26105477
|
0.829
|
NM_001145454
NM_005563
|
STMN1
|
stathmin 1
|
|
chr13_-_109236897
|
0.825
|
NM_003749
|
IRS2
|
insulin receptor substrate 2
|
|
chr1_+_177261627
|
0.825
|
|
FAM20B
|
family with sequence similarity 20, member B
|
|
chr4_-_74154272
|
0.825
|
NM_173827
|
COX18
|
COX18 cytochrome c oxidase assembly homolog (S. cerevisiae)
|
|
chr2_+_153283381
|
0.822
|
|
ARL6IP6
|
ADP-ribosylation-like factor 6 interacting protein 6
|
|
chr5_+_36912595
|
0.818
|
NM_015384
NM_133433
|
NIPBL
|
Nipped-B homolog (Drosophila)
|
|
chr15_+_30694982
|
0.816
|
NM_014783
NM_199357
|
ARHGAP11A
|
Rho GTPase activating protein 11A
|
|
chr15_+_39739933
|
0.815
|
|
MGA
|
MAX gene associated
|
|
chr4_+_120353177
|
0.810
|
|
|
|
|
chr3_+_41215999
|
0.803
|
|
CTNNB1
|
catenin (cadherin-associated protein), beta 1, 88kDa
|
|
chr6_+_27914301
|
0.801
|
NM_003520
|
HIST1H2BN
|
histone cluster 1, H2bn
|
|
chr1_+_61320213
|
0.799
|
NM_001134673
NM_005595
|
NFIA
|
nuclear factor I/A
|
|
chr21_+_29592984
|
0.789
|
|
BACH1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1
|
|
chr14_+_34521945
|
0.784
|
|
SRP54
|
signal recognition particle 54kDa
|
|
chr15_+_39739864
|
0.783
|
NM_001080541
NM_001164273
|
MGA
|
MAX gene associated
|
|
chr2_-_219141199
|
0.781
|
NM_020935
|
USP37
|
ubiquitin specific peptidase 37
|
|
chr1_+_26671488
|
0.779
|
NM_005517
|
HMGN2
|
high-mobility group nucleosomal binding domain 2
|
|
chr5_-_36912487
|
0.779
|
|
LOC646719
|
hypothetical LOC646719
|
|
chr1_-_120413798
|
0.778
|
NM_024408
|
NOTCH2
|
notch 2
|
|
chr6_-_27883662
|
0.777
|
NM_003519
|
HIST1H2BL
|
histone cluster 1, H2bl
|
|
chr7_+_30291136
|
0.774
|
|
ZNRF2
|
zinc and ring finger 2
|
|
chr7_+_90063543
|
0.774
|
|
CDK14
|
cyclin-dependent kinase 14
|
|
chr13_+_78953495
|
0.771
|
|
NDFIP2
|
Nedd4 family interacting protein 2
|
|
chr6_+_108988761
|
0.769
|
NM_001455
|
FOXO3
|
forkhead box O3
|
|
chr13_+_78953367
|
0.763
|
|
NDFIP2
|
Nedd4 family interacting protein 2
|
|
chr11_+_125001063
|
0.763
|
|
CHEK1
|
CHK1 checkpoint homolog (S. pombe)
|
|
chr6_+_135544214
|
0.762
|
|
MYB
|
v-myb myeloblastosis viral oncogene homolog (avian)
|
|
chr4_+_39734902
|
0.757
|
NM_018177
|
N4BP2
|
NEDD4 binding protein 2
|
|
chr22_+_18485068
|
0.753
|
|
RANBP1
|
RAN binding protein 1
|
|
chr6_+_44463265
|
0.750
|
NM_001253
|
CDC5L
|
CDC5 cell division cycle 5-like (S. pombe)
|
|
chr20_-_31737815
|
0.750
|
NM_005225
|
E2F1
|
E2F transcription factor 1
|
|
chr6_-_116681863
|
0.749
|
NM_021648
|
TSPYL4
|
TSPY-like 4
|
|
chr2_+_170149092
|
0.746
|
NM_004792
|
PPIG
|
peptidylprolyl isomerase G (cyclophilin G)
|
|
chr7_-_106088565
|
0.743
|
NM_175884
|
FLJ36031
|
hypothetical protein FLJ36031
|
|
chr6_+_27883877
|
0.741
|
NM_003509
|
HIST1H2AI
HIST1H3F
|
histone cluster 1, H2ai
histone cluster 1, H3f
|
|
chr2_+_153283375
|
0.739
|
|
ARL6IP6
|
ADP-ribosylation-like factor 6 interacting protein 6
|
|
chr21_+_29593088
|
0.731
|
NM_001186
|
BACH1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1
|
|
chr20_+_5879297
|
0.726
|
NM_032485
NM_182802
|
MCM8
|
minichromosome maintenance complex component 8
|
|
chr4_+_120353212
|
0.724
|
NM_019050
|
USP53
|
ubiquitin specific peptidase 53
|
|
chr1_+_109434896
|
0.722
|
NM_020141
|
TMEM167B
|
transmembrane protein 167B
|
|
chr13_+_78953258
|
0.722
|
NM_001161407
NM_019080
|
NDFIP2
|
Nedd4 family interacting protein 2
|
|
chr7_+_30291029
|
0.721
|
|
ZNRF2
|
zinc and ring finger 2
|
|
chr14_+_35365266
|
0.719
|
NM_032352
|
BRMS1L
|
breast cancer metastasis-suppressor 1-like
|
|
chr17_+_40089270
|
0.716
|
NM_001145080
|
C17orf104
|
chromosome 17 open reading frame 104
|
|
chr9_+_99785493
|
0.714
|
|
ANP32B
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member B
|
|
chr2_+_171494026
|
0.711
|
|
GORASP2
|
golgi reassembly stacking protein 2, 55kDa
|
|
chr15_+_54444907
|
0.708
|
NM_198524
|
TEX9
|
testis expressed 9
|
|
chr7_+_94123612
|
0.707
|
NM_001172437
NM_001172438
|
PEG10
|
paternally expressed 10
|
|
chr6_+_64403668
|
0.704
|
|
PHF3
|
PHD finger protein 3
|
|
chr15_+_42507057
|
0.700
|
|
CTDSPL2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2
|
|
chr6_+_64403631
|
0.699
|
|
PHF3
|
PHD finger protein 3
|
|
chr1_+_52380633
|
0.697
|
NM_004799
NM_007323
NM_007324
|
ZFYVE9
|
zinc finger, FYVE domain containing 9
|
|
chr11_+_73981274
|
0.693
|
NM_006591
|
POLD3
|
polymerase (DNA-directed), delta 3, accessory subunit
|
|
chr6_+_64403685
|
0.692
|
|
PHF3
|
PHD finger protein 3
|
|
chr3_+_144203061
|
0.692
|
NM_001080415
|
U2SURP
|
U2 snRNP-associated SURP domain containing
|
|
chr5_-_74098641
|
0.690
|
NM_032380
NM_170681
NM_170691
|
GFM2
|
G elongation factor, mitochondrial 2
|
|
chr2_+_153283353
|
0.684
|
|
ARL6IP6
|
ADP-ribosylation-like factor 6 interacting protein 6
|
|
chr1_-_26105954
|
0.677
|
NM_203399
|
STMN1
|
stathmin 1
|
|
chr1_-_23730298
|
0.675
|
NM_004091
|
E2F2
|
E2F transcription factor 2
|
|
chr7_-_94123362
|
0.674
|
|
SGCE
|
sarcoglycan, epsilon
|
|
chr7_-_94123308
|
0.672
|
|
SGCE
|
sarcoglycan, epsilon
|
|
chr6_-_91353492
|
0.665
|
NM_003188
NM_145332
NM_145333
|
MAP3K7
|
mitogen-activated protein kinase kinase kinase 7
|
|
chr9_+_95253732
|
0.665
|
NM_014612
|
FAM120A
|
family with sequence similarity 120A
|
|
chr14_+_50776635
|
0.664
|
NM_030755
|
TMX1
|
thioredoxin-related transmembrane protein 1
|
|
chr1_+_28029922
|
0.660
|
|
PPP1R8
|
protein phosphatase 1, regulatory (inhibitor) subunit 8
|
|
chr9_-_103289154
|
0.660
|
NM_032342
|
C9orf125
|
chromosome 9 open reading frame 125
|
|
chr1_-_120413783
|
0.656
|
|
NOTCH2
|
notch 2
|
|
chr1_+_62675323
|
0.651
|
|
USP1
|
ubiquitin specific peptidase 1
|
|
chr4_+_148758014
|
0.650
|
|
TMEM184C
|
transmembrane protein 184C
|
|
chr21_+_29593098
|
0.650
|
|
BACH1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1
|
|
chr7_-_94123413
|
0.649
|
NM_001099400
NM_001099401
NM_003919
|
SGCE
|
sarcoglycan, epsilon
|
|
chr3_+_113180517
|
0.646
|
NM_018394
|
ABHD10
|
abhydrolase domain containing 10
|
|
chr6_+_139497933
|
0.645
|
NM_016217
|
HECA
|
headcase homolog (Drosophila)
|
|
chr1_+_28029909
|
0.640
|
|
PPP1R8
|
protein phosphatase 1, regulatory (inhibitor) subunit 8
|
|
chr10_+_13243553
|
0.639
|
NM_018518
NM_182751
|
MCM10
|
minichromosome maintenance complex component 10
|
|
chr7_+_94123556
|
0.638
|
NM_001040152
NM_001184961
NM_001184962
NM_015068
|
PEG10
|
paternally expressed 10
|
|
chr7_-_77163475
|
0.632
|
|
|
|
|
chr12_+_70519753
|
0.629
|
NM_001146213
NM_001146214
NM_022771
|
TBC1D15
|
TBC1 domain family, member 15
|
|
chr1_+_28029879
|
0.628
|
NM_002713
NM_014110
NM_138558
|
PPP1R8
|
protein phosphatase 1, regulatory (inhibitor) subunit 8
|
|
chr1_+_205992032
|
0.627
|
|
CD46
|
CD46 molecule, complement regulatory protein
|
|
chr4_+_152240182
|
0.626
|
NM_001006
|
RPS3A
|
ribosomal protein S3A
|
|
chr5_+_75414994
|
0.624
|
NM_014979
|
SV2C
|
synaptic vesicle glycoprotein 2C
|
|
chr1_+_62675296
|
0.624
|
|
USP1
|
ubiquitin specific peptidase 1
|
|
chr1_-_120413698
|
0.623
|
|
NOTCH2
|
notch 2
|
|
chr1_-_91260181
|
0.623
|
NM_032186
NM_201269
|
ZNF644
|
zinc finger protein 644
|
|
chr15_+_42507123
|
0.621
|
|
CTDSPL2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2
|
|
chr1_-_39097904
|
0.620
|
NM_022157
|
RRAGC
|
Ras-related GTP binding C
|
|
chr17_+_26082779
|
0.619
|
|
SUZ12P
|
suppressor of zeste 12 homolog pseudogene
|
|
chr8_+_21833162
|
0.615
|
|
XPO7
|
exportin 7
|
|
chr22_+_18485145
|
0.614
|
|
RANBP1
|
RAN binding protein 1
|
|
chr6_+_64403936
|
0.613
|
|
PHF3
|
PHD finger protein 3
|
|
chr6_+_157141565
|
0.611
|
|
ARID1B
|
AT rich interactive domain 1B (SWI1-like)
|
|
chr4_+_148757958
|
0.610
|
NM_018241
|
TMEM184C
|
transmembrane protein 184C
|
|
chr7_-_151764004
|
0.606
|
NM_170606
|
MLL3
|
myeloid/lymphoid or mixed-lineage leukemia 3
|
|
chr6_+_163755773
|
0.606
|
|
QKI
|
quaking homolog, KH domain RNA binding (mouse)
|
|
chr2_+_64605049
|
0.595
|
|
AFTPH
|
aftiphilin
|
|
chr5_+_72287509
|
0.595
|
NM_001146032
NM_138782
|
FCHO2
|
FCH domain only 2
|
|
chr5_+_138657317
|
0.594
|
NM_001194955
NM_001194956
|
MATR3
|
matrin 3
|
|
chr20_-_48980858
|
0.592
|
NM_015339
NM_181442
|
ADNP
|
activity-dependent neuroprotector homeobox
|
|
chr11_+_9552005
|
0.590
|
|
WEE1
|
WEE1 homolog (S. pombe)
|
|
chr11_+_9551885
|
0.589
|
|
WEE1
|
WEE1 homolog (S. pombe)
|
|
chr5_+_138657229
|
0.588
|
NM_018834
|
MATR3
|
matrin 3
|
|
chrX_+_130985316
|
0.587
|
NM_001042452
|
MST4
|
serine/threonine protein kinase MST4
|
|
chr9_+_85785456
|
0.587
|
NM_024945
|
RMI1
|
RMI1, RecQ mediated genome instability 1, homolog (S. cerevisiae)
|
|
chr14_+_34521854
|
0.579
|
NM_001146282
NM_003136
|
SRP54
|
signal recognition particle 54kDa
|
|
chr1_+_100371293
|
0.579
|
NM_019083
|
CCDC76
|
coiled-coil domain containing 76
|
|
chr1_-_100370994
|
0.574
|
NM_194292
|
SASS6
|
spindle assembly 6 homolog (C. elegans)
|
|
chr8_+_21833111
|
0.574
|
NM_015024
|
XPO7
|
exportin 7
|
|
chr16_+_71256497
|
0.573
|
|
|
|
|
chr20_-_21326046
|
0.573
|
NM_033176
|
NKX2-4
|
NK2 homeobox 4
|
|
chr15_+_66133625
|
0.571
|
NM_016166
|
PIAS1
|
protein inhibitor of activated STAT, 1
|
|
chr20_+_36095263
|
0.570
|
NM_021215
|
RPRD1B
|
regulation of nuclear pre-mRNA domain containing 1B
|
|
chr6_+_27969181
|
0.570
|
NM_003527
|
HIST1H2BO
|
histone cluster 1, H2bo
|
|
chr9_-_122679387
|
0.569
|
NM_001009936
NM_015651
|
PHF19
|
PHD finger protein 19
|
|
chr5_+_72287625
|
0.569
|
|
|
|