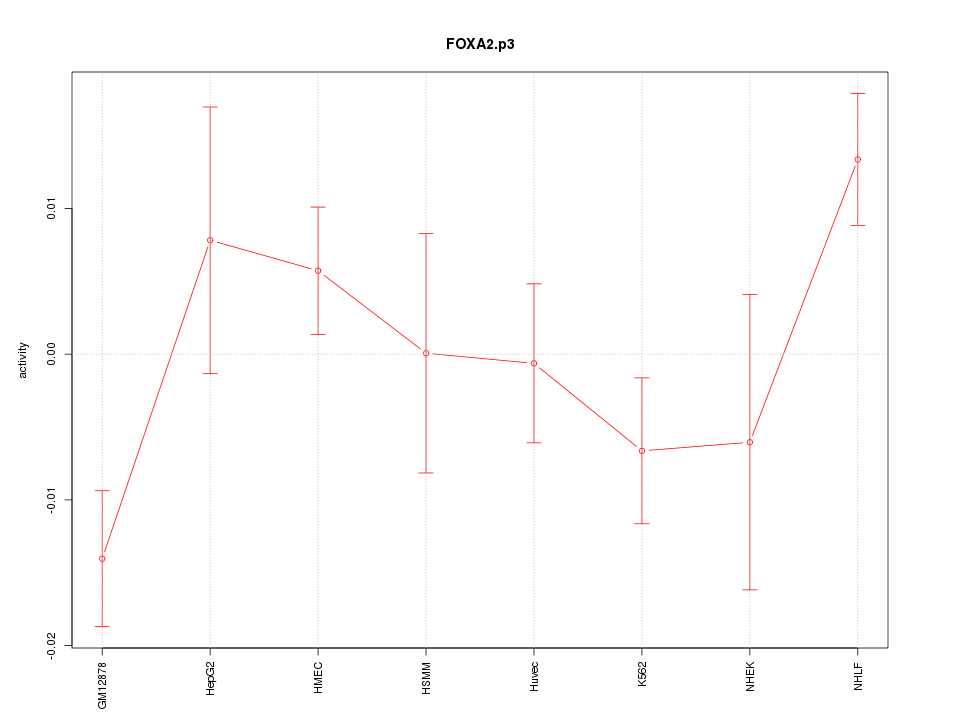
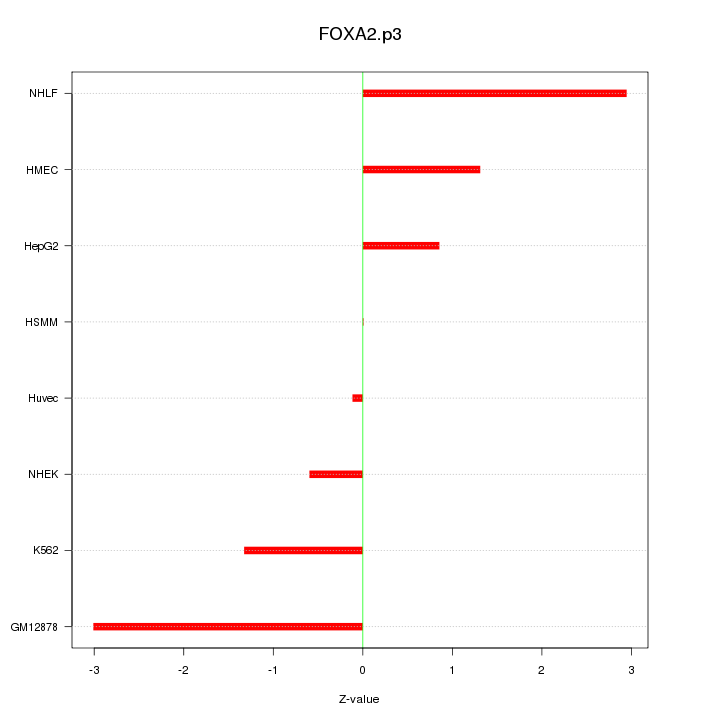
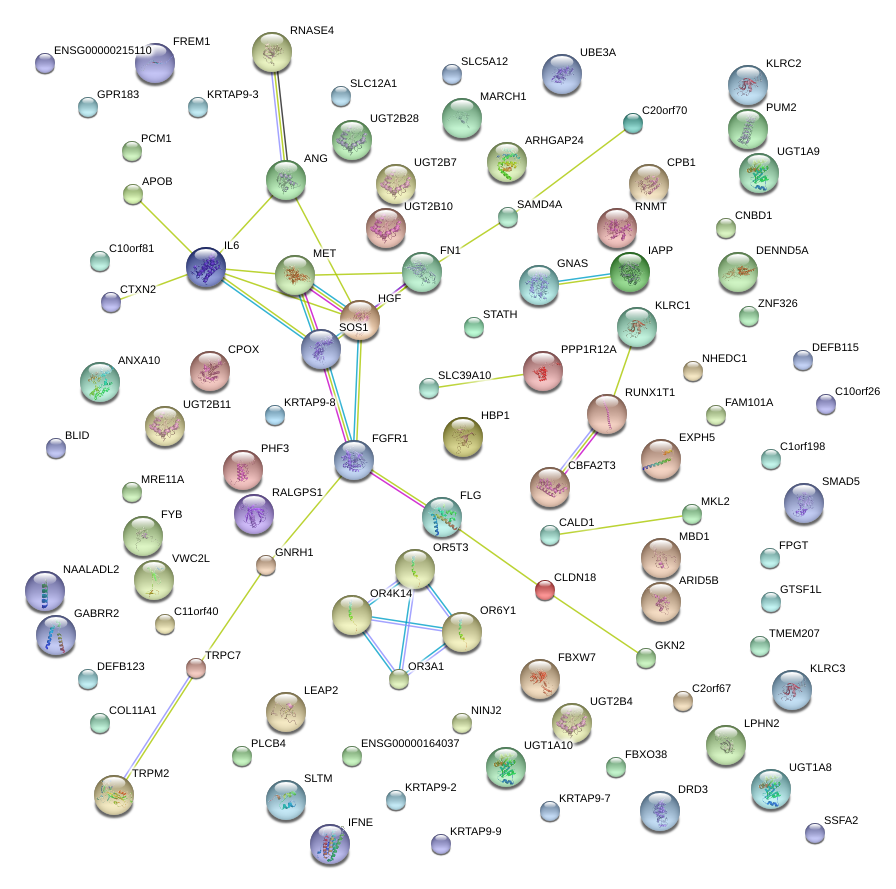
|
chr15_-_23204887
|
3.734
|
NM_000462
|
UBE3A
|
ubiquitin protein ligase E3A
|
|
chr2_-_55053398
|
2.373
|
|
|
|
|
chr4_+_69716298
|
2.267
|
NM_001075
NM_001144767
|
UGT2B10
|
UDP glucuronosyltransferase 2 family, polypeptide B10
|
|
chr17_+_36642240
|
1.738
|
NM_031962
|
KRTAP9-9
KRTAP9-3
|
keratin associated protein 9-9
keratin associated protein 9-3
|
|
chr12_+_21417068
|
1.651
|
NM_000415
|
IAPP
|
islet amyloid polypeptide
|
|
chr10_+_63478974
|
1.594
|
|
ARID5B
|
AT rich interactive domain 5B (MRF1-like)
|
|
chr2_-_215967442
|
1.584
|
|
FN1
|
fibronectin 1
|
|
chr2_-_21079145
|
1.496
|
|
APOB
|
apolipoprotein B (including Ag(x) antigen)
|
|
chr20_+_9406460
|
1.496
|
|
PLCB4
|
phospholipase C, beta 4
|
|
chr8_-_25338472
|
1.472
|
NM_000825
NM_001083111
|
GNRH1
|
gonadotropin-releasing hormone 1 (luteinizing-releasing hormone)
|
|
chr5_+_147754467
|
1.464
|
|
FBXO38
|
F-box protein 38
|
|
chr4_-_70115037
|
1.418
|
NM_001073
|
UGT2B11
|
UDP glucuronosyltransferase 2 family, polypeptide B11
|
|
chr1_-_103269356
|
1.417
|
|
COL11A1
|
collagen, type XI, alpha 1
|
|
chr2_-_21084911
|
1.395
|
|
APOB
|
apolipoprotein B (including Ag(x) antigen)
|
|
chr14_+_54238581
|
1.350
|
|
SAMD4A
|
sterile alpha motif domain containing 4A
|
|
chr2_-_210726539
|
1.350
|
|
C2orf67
|
chromosome 2 open reading frame 67
|
|
chr3_-_121065112
|
1.348
|
|
|
|
|
chr1_-_156784518
|
1.305
|
NM_001005189
|
OR6Y1
|
olfactory receptor, family 6, subfamily Y, member 1
|
|
chr17_+_36665161
|
1.269
|
NM_030975
|
KRTAP9-9
|
keratin associated protein 9-9
|
|
chr6_+_64414389
|
1.234
|
NM_015153
|
PHF3
|
PHD finger protein 3
|
|
chr6_+_64403668
|
1.233
|
|
PHF3
|
PHD finger protein 3
|
|
chr6_+_131190237
|
1.232
|
NM_001195597
|
LOC285733
|
hypothetical LOC285733
|
|
chr4_+_169250281
|
1.220
|
NM_007193
|
ANXA10
|
annexin A10
|
|
chr3_-_115380539
|
1.204
|
NM_000796
NM_033663
|
DRD3
|
dopamine receptor D3
|
|
chr2_+_182464910
|
1.203
|
|
SSFA2
|
sperm specific antigen 2
|
|
chr8_+_87947788
|
1.194
|
NM_173538
|
CNBD1
|
cyclic nucleotide binding domain containing 1
|
|
chr1_+_74473672
|
1.191
|
NM_015978
|
TNNI3K
|
TNNI3 interacting kinase
|
|
chr7_+_134226727
|
1.186
|
|
CALD1
|
caldesmon 1
|
|
chr2_-_21084077
|
1.184
|
|
APOB
|
apolipoprotein B (including Ag(x) antigen)
|
|
chr20_+_31219543
|
1.168
|
NM_080574
|
C20orf70
|
chromosome 20 open reading frame 70
|
|
chr12_-_10479810
|
1.165
|
NM_002260
|
KLRC2
KLRC3
|
killer cell lectin-like receptor subfamily C, member 2
killer cell lectin-like receptor subfamily C, member 3
|
|
chr5_+_132237253
|
1.152
|
NM_052971
|
LEAP2
|
liver expressed antimicrobial peptide 2
|
|
chr11_-_26700121
|
1.149
|
NM_178498
|
SLC5A12
|
solute carrier family 5 (sodium/glucose cotransporter), member 12
|
|
chr5_+_10406980
|
1.128
|
|
|
|
|
chr11_+_55776251
|
1.128
|
NM_001004747
|
OR5T3
|
olfactory receptor, family 5, subfamily T, member 3
|
|
chr7_+_106596641
|
1.124
|
NM_012257
|
HBP1
|
HMG-box transcription factor 1
|
|
chr20_+_56912146
|
1.120
|
|
GNAS
|
GNAS complex locus
|
|
chr5_+_135496421
|
1.117
|
NM_001001419
NM_001001420
NM_005903
|
SMAD5
|
SMAD family member 5
|
|
chr11_-_121492010
|
1.086
|
NM_001001786
|
BLID
|
BH3-like motif containing, cell death inducer
|
|
chr3_+_150028130
|
1.061
|
NM_001871
|
CPB1
|
carboxypeptidase B1 (tissue)
|
|
chr1_+_81544432
|
1.041
|
|
LPHN2
|
latrophilin 2
|
|
chr3_+_70131572
|
1.030
|
|
|
|
|
chr12_-_642786
|
0.982
|
NM_016533
|
NINJ2
|
ninjurin 2
|
|
chr20_-_41788975
|
0.957
|
NM_001008901
NM_176791
|
GTSF1L
|
gametocyte specific factor 1-like
|
|
chr2_+_196229715
|
0.947
|
NM_001127257
|
SLC39A10
|
solute carrier family 39 (zinc transporter), member 10
|
|
chr3_+_139200347
|
0.938
|
NM_001002026
|
CLDN18
|
claudin 18
|
|
chr11_-_107969554
|
0.936
|
NM_015065
|
EXPH5
|
exophilin 5
|
|
chr2_+_214984705
|
0.933
|
NM_001080500
|
VWC2L
|
von Willebrand factor C domain containing protein 2-like
|
|
chr4_+_70896236
|
0.932
|
NM_001009181
NM_003154
|
STATH
|
statherin
|
|
chr5_-_39238818
|
0.929
|
|
FYB
|
FYN binding protein
|
|
chr9_-_21472311
|
0.923
|
NM_176891
|
IFNE
|
interferon, epsilon
|
|
chr12_-_10464308
|
0.920
|
NM_002261
NM_007333
|
KLRC3
|
killer cell lectin-like receptor subfamily C, member 3
|
|
chr5_+_135496437
|
0.916
|
|
SMAD5
|
SMAD family member 5
|
|
chr2_+_196230096
|
0.912
|
NM_020342
|
SLC39A10
|
solute carrier family 39 (zinc transporter), member 10
|
|
chr13_-_98757634
|
0.894
|
NM_004951
|
GPR183
|
G protein-coupled receptor 183
|
|
chr11_-_4555625
|
0.894
|
NM_144663
|
C11orf40
|
chromosome 11 open reading frame 40
|
|
chr8_+_17866373
|
0.882
|
|
PCM1
|
pericentriolar material 1
|
|
chr2_-_21081694
|
0.861
|
|
APOB
|
apolipoprotein B (including Ag(x) antigen)
|
|
chr9_-_14900992
|
0.856
|
NM_144966
|
FREM1
|
FRAS1 related extracellular matrix 1
|
|
chr8_+_17874128
|
0.851
|
|
PCM1
|
pericentriolar material 1
|
|
chr14_+_20226762
|
0.848
|
NM_001097577
NM_194431
|
ANG
RNASE4
|
angiogenin, ribonuclease, RNase A family, 5
ribonuclease, RNase A family, 4
|
|
chr3_+_176059763
|
0.844
|
NM_207015
|
NAALADL2
|
N-acetylated alpha-linked acidic dipeptidase-like 2
|
|
chr16_+_14187965
|
0.842
|
|
MKL2
|
MKL/myocardin-like 2
|
|
chr10_+_104525877
|
0.840
|
NM_017787
|
C10orf26
|
chromosome 10 open reading frame 26
|
|
chr2_-_20390601
|
0.835
|
NM_015317
|
PUM2
|
pumilio homolog 2 (Drosophila)
|
|
chr20_+_29492071
|
0.826
|
NM_153324
|
DEFB123
|
defensin, beta 123
|
|
chr4_+_86615275
|
0.825
|
NM_001025616
|
ARHGAP24
|
Rho GTPase activating protein 24
|
|
chr7_+_116151411
|
0.824
|
|
MET
|
met proto-oncogene (hepatocyte growth factor receptor)
|
|
chr15_+_46271158
|
0.822
|
NM_001145668
|
CTXN2
SLC12A1
|
cortexin 2
solute carrier family 12 (sodium/potassium/chloride transporters), member 1
|
|
chr4_-_164754106
|
0.811
|
NM_017923
|
MARCH1
|
membrane-associated ring finger (C3HC4) 1
|
|
chr14_-_19553191
|
0.809
|
NM_001004712
|
OR4K14
|
olfactory receptor, family 4, subfamily K, member 14
|
|
chr5_+_27508139
|
0.794
|
|
LOC643401
|
hypothetical protein LOC643401
|
|
chr17_-_3142625
|
0.774
|
NM_002550
|
OR3A1
|
olfactory receptor, family 3, subfamily A, member 1
|
|
chr2_-_69033515
|
0.771
|
NM_182536
|
GKN2
|
gastrokine 2
|
|
chr16_-_87570716
|
0.759
|
NM_005187
|
CBFA2T3
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 3
|
|
chr1_+_90233238
|
0.757
|
NM_182975
NM_182976
|
ZNF326
|
zinc finger protein 326
|
|
chr3_-_99795074
|
0.749
|
NM_000097
|
CPOX
|
coproporphyrinogen oxidase
|
|
chr1_-_150564302
|
0.740
|
NM_002016
|
FLG
|
filaggrin
|
|
chr11_-_9182356
|
0.732
|
|
DENND5A
|
DENN/MADD domain containing 5A
|
|
chr9_+_128764324
|
0.731
|
NM_001190728
|
RALGPS1
|
Ral GEF with PH domain and SH3 binding motif 1
|
|
chr2_+_234191029
|
0.726
|
NM_019076
|
UGT1A8
|
UDP glucuronosyltransferase 1 family, polypeptide A8
|
|
chr4_-_104160291
|
0.725
|
NM_001100874
NM_139173
|
NHEDC1
|
Na+/H+ exchanger domain containing 1
|
|
chr12_-_78831821
|
0.722
|
NM_001143886
|
PPP1R12A
|
protein phosphatase 1, regulatory (inhibitor) subunit 12A
|
|
chr3_-_191650291
|
0.721
|
NM_207316
|
TMEM207
|
transmembrane protein 207
|
|
chr20_+_29309127
|
0.715
|
NM_001037730
|
DEFB115
|
defensin, beta 115
|
|
chr1_-_229058404
|
0.711
|
NM_001136495
|
C1orf198
|
chromosome 1 open reading frame 198
|
|
chr6_-_90081655
|
0.710
|
NM_002043
|
GABRR2
|
gamma-aminobutyric acid (GABA) receptor, rho 2
|
|
chr12_+_123339662
|
0.703
|
NM_181709
|
FAM101A
|
family with sequence similarity 101, member A
|
|
chr4_-_153523113
|
0.701
|
NM_001013415
|
FBXW7
|
F-box and WD repeat domain containing 7
|
|
chr5_-_135729062
|
0.700
|
NM_001167576
NM_001167577
NM_020389
|
TRPC7
|
transient receptor potential cation channel, subfamily C, member 7
|
|
chr15_+_22781292
|
0.697
|
|
PAR5
|
Prader-Willi/Angelman syndrome-5
|
|
chr2_-_65013070
|
0.691
|
|
LOC400958
|
hypothetical LOC400958
|
|
chr8_-_93099040
|
0.690
|
NM_175636
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr11_-_93866554
|
0.684
|
NM_005590
NM_005591
|
MRE11A
|
MRE11 meiotic recombination 11 homolog A (S. cerevisiae)
|
|
chr7_-_81237162
|
0.683
|
NM_000601
NM_001010931
NM_001010932
NM_001010933
NM_001010934
|
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor)
|
|
chr4_+_70180805
|
0.681
|
NM_053039
|
UGT2B28
|
UDP glucuronosyltransferase 2 family, polypeptide B28
|
|
chr1_+_86662356
|
0.680
|
NM_006536
|
CLCA2
|
chloride channel accessory 2
|
|
chr6_+_127940011
|
0.678
|
NM_001010905
|
C6orf58
|
chromosome 6 open reading frame 58
|
|
chr3_+_99334230
|
0.677
|
NM_001005338
|
OR5H1
|
olfactory receptor, family 5, subfamily H, member 1
|
|
chr5_+_52892217
|
0.677
|
NM_002495
|
NDUFS4
|
NADH dehydrogenase (ubiquinone) Fe-S protein 4, 18kDa (NADH-coenzyme Q reductase)
|
|
chr4_-_140443061
|
0.677
|
NM_001184987
NM_001184988
NM_001184989
NM_001184990
|
NDUFC1
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa
|
|
chr3_+_50687675
|
0.674
|
NM_004947
|
DOCK3
|
dedicator of cytokinesis 3
|
|
chr4_-_164614335
|
0.673
|
NM_032136
|
TKTL2
|
transketolase-like 2
|
|
chr3_+_179759181
|
0.672
|
NM_005832
|
KCNMB2
|
potassium large conductance calcium-activated channel, subfamily M, beta member 2
|
|
chr2_+_170149348
|
0.670
|
|
PPIG
|
peptidylprolyl isomerase G (cyclophilin G)
|
|
chr18_+_64616296
|
0.670
|
NM_024781
|
CCDC102B
|
coiled-coil domain containing 102B
|
|
chr4_+_157044296
|
0.668
|
NM_005651
|
TDO2
|
tryptophan 2,3-dioxygenase
|
|
chr14_+_36200815
|
0.667
|
|
PAX9
|
paired box 9
|
|
chr10_-_49152918
|
0.665
|
NM_001018071
|
FRMPD2
|
FERM and PDZ domain containing 2
|
|
chr7_+_90177082
|
0.663
|
|
CDK14
|
cyclin-dependent kinase 14
|
|
chr6_+_7052985
|
0.659
|
NM_001003698
NM_001003699
NM_001003700
|
RREB1
|
ras responsive element binding protein 1
|
|
chr10_+_124729899
|
0.658
|
|
PSTK
|
phosphoseryl-tRNA kinase
|
|
chr3_-_114176401
|
0.656
|
NM_138806
NM_138939
NM_138940
NM_170780
|
CD200R1
|
CD200 receptor 1
|
|
chr14_-_105644300
|
0.647
|
|
IGHV4-31
IGHA1
IGHA2
IGHG1
IGHG3
IGHM
|
immunoglobulin heavy variable 4-31
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant alpha 2 (A2m marker)
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant gamma 3 (G3m marker)
immunoglobulin heavy constant mu
|
|
chr12_-_11066436
|
0.641
|
NM_176888
|
TAS2R19
|
taste receptor, type 2, member 19
|
|
chr9_-_34700062
|
0.641
|
NM_002989
|
CCL21
|
chemokine (C-C motif) ligand 21
|
|
chr11_+_55842358
|
0.633
|
NM_001005202
|
OR8K3
|
olfactory receptor, family 8, subfamily K, member 3
|
|
chr9_+_6403321
|
0.632
|
|
UHRF2
|
ubiquitin-like with PHD and ring finger domains 2
|
|
chr6_-_127822227
|
0.632
|
NM_014702
|
KIAA0408
|
KIAA0408
|
|
chr1_-_150599105
|
0.631
|
NM_001014342
|
FLG2
|
filaggrin family member 2
|
|
chr18_+_28023984
|
0.627
|
NM_005925
|
MEP1B
|
meprin A, beta
|
|
chr6_+_7053183
|
0.626
|
|
RREB1
|
ras responsive element binding protein 1
|
|
chr3_+_173951163
|
0.624
|
|
ECT2
|
epithelial cell transforming sequence 2 oncogene
|
|
chr12_+_63090008
|
0.622
|
|
XPOT
|
exportin, tRNA (nuclear export receptor for tRNAs)
|
|
chr12_+_99391870
|
0.619
|
|
NR1H4
|
nuclear receptor subfamily 1, group H, member 4
|
|
chr11_-_27637771
|
0.619
|
NM_001143816
NM_170735
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr12_-_11041740
|
0.615
|
NM_176889
|
TAS2R20
|
taste receptor, type 2, member 20
|
|
chr13_+_75232797
|
0.615
|
NM_015842
|
LMO7
|
LIM domain 7
|
|
chr14_+_55654900
|
0.614
|
|
PELI2
|
pellino homolog 2 (Drosophila)
|
|
chr6_+_154373323
|
0.605
|
NM_001145279
NM_001145280
NM_001145281
|
OPRM1
|
opioid receptor, mu 1
|
|
chr6_+_55147029
|
0.602
|
NM_001526
|
HCRTR2
|
hypocretin (orexin) receptor 2
|
|
chr5_+_52892229
|
0.601
|
|
NDUFS4
|
NADH dehydrogenase (ubiquinone) Fe-S protein 4, 18kDa (NADH-coenzyme Q reductase)
|
|
chr4_+_69465107
|
0.601
|
NM_014058
|
TMPRSS11E
|
transmembrane protease, serine 11E
|
|
chr5_+_81636921
|
0.600
|
NM_001017971
|
ATP6AP1L
|
ATPase, H+ transporting, lysosomal accessory protein 1-like
|
|
chr3_-_87408301
|
0.598
|
NM_000306
NM_001122757
|
POU1F1
|
POU class 1 homeobox 1
|
|
chr6_-_47030492
|
0.595
|
NM_015234
|
GPR116
|
G protein-coupled receptor 116
|
|
chr3_-_194755281
|
0.593
|
NM_032279
|
ATP13A4
|
ATPase type 13A4
|
|
chr2_+_191009265
|
0.593
|
|
MFSD6
|
major facilitator superfamily domain containing 6
|
|
chr14_+_36200867
|
0.593
|
|
PAX9
|
paired box 9
|
|
chr9_+_6403183
|
0.582
|
|
UHRF2
|
ubiquitin-like with PHD and ring finger domains 2
|
|
chr18_+_27425727
|
0.582
|
NM_000371
|
TTR
|
transthyretin
|
|
chrX_+_24077782
|
0.579
|
|
ZFX
|
zinc finger protein, X-linked
|
|
chr7_-_132371208
|
0.578
|
|
EEF1G
|
eukaryotic translation elongation factor 1 gamma
|
|
chr2_+_183316574
|
0.577
|
|
DNAJC10
|
DnaJ (Hsp40) homolog, subfamily C, member 10
|
|
chr14_+_18447521
|
0.573
|
NM_001013354
|
OR11H12
|
olfactory receptor, family 11, subfamily H, member 12
|
|
chrX_+_24077649
|
0.571
|
NM_001178086
NM_001178095
NM_001178084
NM_001178085
NM_003410
|
ZFX
|
zinc finger protein, X-linked
|
|
chr4_+_35959638
|
0.571
|
NM_001136536
|
DTHD1
|
death domain containing 1
|
|
chr5_-_58607673
|
0.570
|
NM_001197220
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr8_+_21833111
|
0.567
|
NM_015024
|
XPO7
|
exportin 7
|
|
chr1_+_69998445
|
0.567
|
NM_020794
|
LRRC7
|
leucine rich repeat containing 7
|
|
chr17_+_64922420
|
0.567
|
NM_002758
|
MAP2K6
|
mitogen-activated protein kinase kinase 6
|
|
chr10_+_124729545
|
0.565
|
NM_153336
|
PSTK
|
phosphoseryl-tRNA kinase
|
|
chr5_+_147775652
|
0.565
|
|
FBXO38
|
F-box protein 38
|
|
chr5_-_13997588
|
0.561
|
NM_001369
|
DNAH5
|
dynein, axonemal, heavy chain 5
|
|
chr3_+_173954940
|
0.558
|
NM_018098
|
ECT2
|
epithelial cell transforming sequence 2 oncogene
|
|
chr4_+_111616630
|
0.556
|
NM_001977
|
ENPEP
|
glutamyl aminopeptidase (aminopeptidase A)
|
|
chr7_+_111879348
|
0.555
|
NM_001197079
|
IFRD1
|
interferon-related developmental regulator 1
|
|
chr4_+_71054492
|
0.554
|
NM_214711
|
C4orf40
|
chromosome 4 open reading frame 40
|
|
chr7_+_80069439
|
0.550
|
NM_001001547
|
CD36
|
CD36 molecule (thrombospondin receptor)
|
|
chr2_+_190248955
|
0.547
|
NM_144708
|
ANKAR
|
ankyrin and armadillo repeat containing
|
|
chr5_-_43326922
|
0.547
|
|
HMGCS1
|
3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble)
|
|
chr3_-_150569574
|
0.544
|
|
TM4SF1
|
transmembrane 4 L six family member 1
|
|
chr7_+_134226690
|
0.542
|
NM_033139
NM_033140
|
CALD1
|
caldesmon 1
|
|
chr6_+_132933153
|
0.540
|
NM_175067
|
TAAR6
|
trace amine associated receptor 6
|
|
chr2_-_99412694
|
0.537
|
|
REV1
|
REV1 homolog (S. cerevisiae)
|
|
chr17_-_46296376
|
0.537
|
NM_005749
|
TOB1
|
transducer of ERBB2, 1
|
|
chr8_+_21833162
|
0.536
|
|
XPO7
|
exportin 7
|
|
chr20_-_52120421
|
0.535
|
|
BCAS1
|
breast carcinoma amplified sequence 1
|
|
chr2_-_71307705
|
0.534
|
NM_020459
|
PAIP2B
|
poly(A) binding protein interacting protein 2B
|
|
chr12_-_14929991
|
0.532
|
NM_000900
NM_001190839
|
MGP
|
matrix Gla protein
|
|
chr13_-_47916803
|
0.528
|
NM_005767
|
LPAR6
|
lysophosphatidic acid receptor 6
|
|
chr14_+_64041170
|
0.525
|
|
ZBTB1
|
zinc finger and BTB domain containing 1
|
|
chr11_-_55128424
|
0.525
|
NM_001004700
|
OR4C11
|
olfactory receptor, family 4, subfamily C, member 11
|
|
chr10_-_129581192
|
0.523
|
NM_152311
|
CLRN3
|
clarin 3
|
|
chr3_+_35696119
|
0.522
|
NM_001025068
NM_001025069
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa
|
|
chr14_-_24173205
|
0.520
|
NM_004131
|
GZMB
|
granzyme B (granzyme 2, cytotoxic T-lymphocyte-associated serine esterase 1)
|
|
chr1_+_246578699
|
0.519
|
NM_001001918
|
OR14C36
|
olfactory receptor, family 14, subfamily C, member 36
|
|
chr3_+_186563529
|
0.515
|
NM_004721
|
MAP3K13
|
mitogen-activated protein kinase kinase kinase 13
|
|
chr6_+_118976134
|
0.515
|
NM_002667
|
PLN
|
phospholamban
|
|
chr12_+_6745947
|
0.514
|
|
PTMS
|
parathymosin
|
|
chrX_+_123307812
|
0.514
|
NM_001114937
NM_002351
|
SH2D1A
|
SH2 domain containing 1A
|
|
chr21_-_38955487
|
0.511
|
NM_001136154
NM_004449
|
ERG
|
v-ets erythroblastosis virus E26 oncogene homolog (avian)
|
|
chr7_+_134483305
|
0.511
|
NM_018295
|
TMEM140
|
transmembrane protein 140
|
|
chr2_+_211129567
|
0.504
|
NM_001875
|
CPS1
|
carbamoyl-phosphate synthase 1, mitochondrial
|
|
chr11_+_22646229
|
0.499
|
NM_177553
|
GAS2
|
growth arrest-specific 2
|
|
chr2_+_170149092
|
0.495
|
NM_004792
|
PPIG
|
peptidylprolyl isomerase G (cyclophilin G)
|
|
chr3_+_23904086
|
0.493
|
|
UBE2E1
|
ubiquitin-conjugating enzyme E2E 1 (UBC4/5 homolog, yeast)
|
|
chr3_+_145174856
|
0.492
|
NM_001134470
|
C3orf58
|
chromosome 3 open reading frame 58
|
|
chr7_+_94377145
|
0.492
|
NM_001166161
NM_001166162
NM_001166163
|
PPP1R9A
|
protein phosphatase 1, regulatory (inhibitor) subunit 9A
|
|
chr10_-_54201400
|
0.492
|
NM_000242
|
MBL2
|
mannose-binding lectin (protein C) 2, soluble
|
|
chr10_+_90336489
|
0.490
|
NM_001010939
|
LIPJ
|
lipase, family member J
|
|
chr6_-_50045296
|
0.490
|
NM_001037729
|
DEFB113
|
defensin, beta 113
|
|
chr3_+_99370233
|
0.487
|
NM_001005515
|
OR5H15
|
olfactory receptor, family 5, subfamily H, member 15
|
|
chr11_-_5178505
|
0.485
|
NM_001004760
|
OR51V1
|
olfactory receptor, family 51, subfamily V, member 1
|
|
chr17_-_36276987
|
0.481
|
NM_000223
|
KRT12
|
keratin 12
|
|
chr10_-_75005421
|
0.480
|
NM_152586
|
USP54
|
ubiquitin specific peptidase 54
|
|
chr4_-_152368492
|
0.479
|
|
SH3D19
|
SH3 domain containing 19
|
|
chr2_-_36929520
|
0.476
|
|
STRN
|
striatin, calmodulin binding protein
|
|
chr14_+_87560646
|
0.475
|
|
LOC283587
|
hypothetical protein LOC283587
|
|
chr1_-_214663360
|
0.474
|
NM_007123
NM_206933
|
USH2A
|
Usher syndrome 2A (autosomal recessive, mild)
|