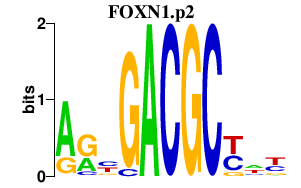
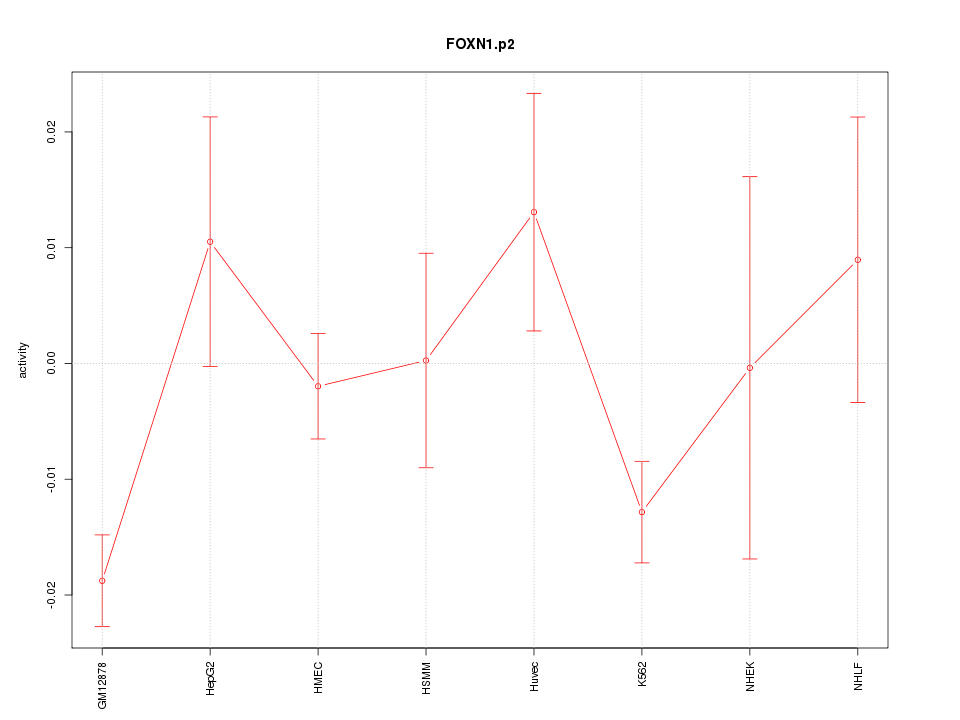
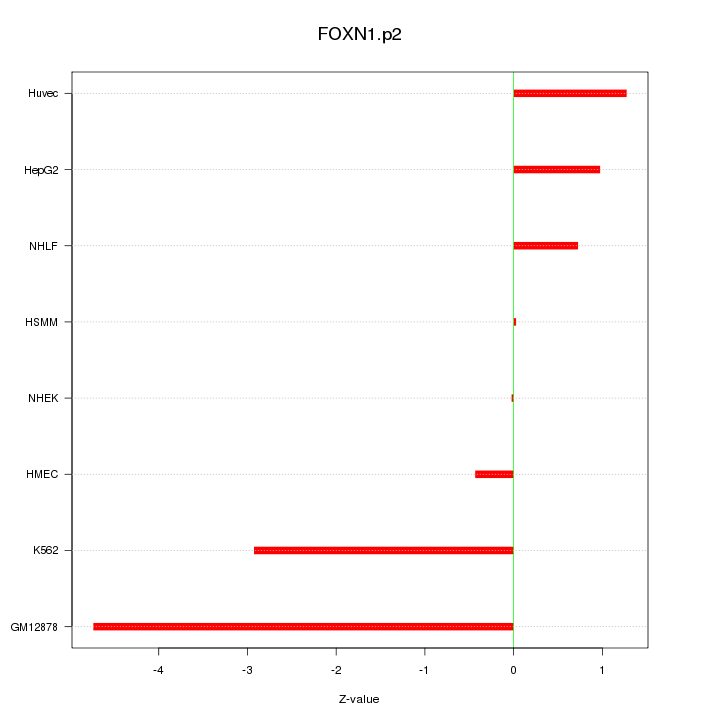
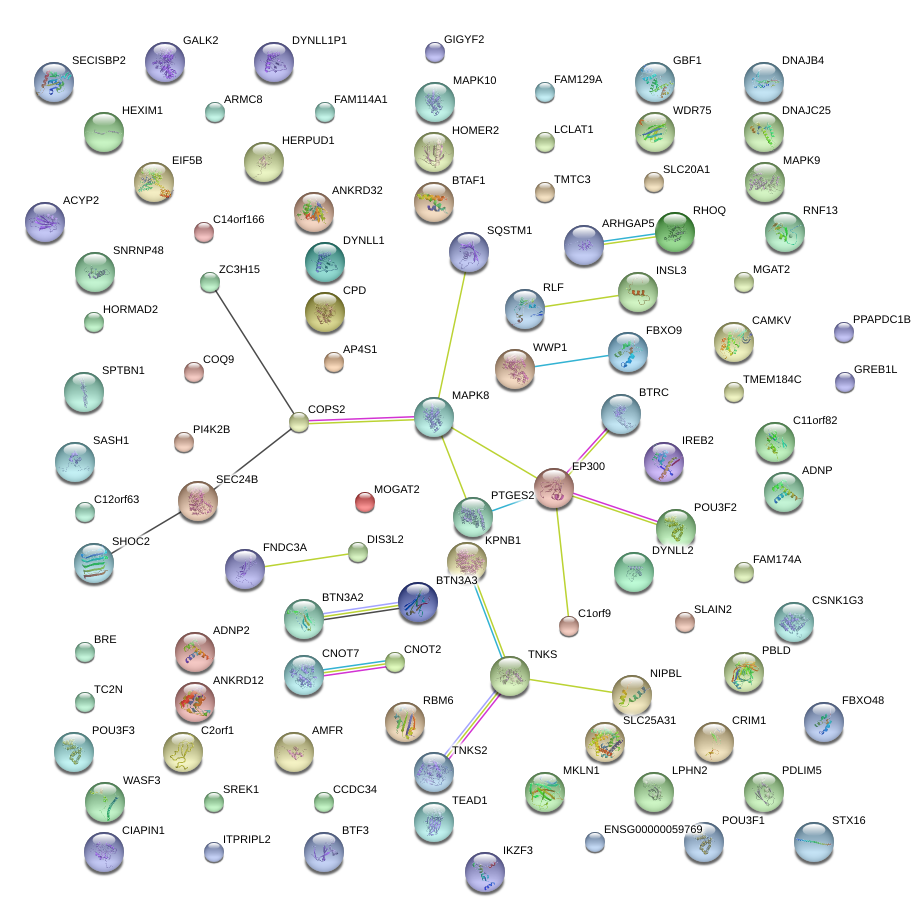
|
chr5_+_122875691
|
3.273
|
NM_001031812
NM_001044722
NM_001044723
NM_004384
|
CSNK1G3
|
casein kinase 1, gamma 3
|
|
chr2_+_30523586
|
3.007
|
NM_182551
NM_001002257
|
LCLAT1
|
lysocardiolipin acyltransferase 1
|
|
chr8_+_87424074
|
2.916
|
NM_007013
|
WWP1
|
WW domain containing E3 ubiquitin protein ligase 1
|
|
chr4_+_119419354
|
2.830
|
|
SNHG8
|
small nucleolar RNA host gene 8 (non-protein coding)
|
|
chr1_+_82038669
|
2.709
|
NM_012302
|
LPHN2
|
latrophilin 2
|
|
chr3_+_139388770
|
2.494
|
NM_014154
NM_015396
NM_213654
|
ARMC8
|
armadillo repeat containing 8
|
|
chr9_+_113433364
|
2.477
|
NM_001015882
NM_004125
|
DNAJC25
DNAJC25-GNG10
|
DnaJ (Hsp40) homolog, subfamily C , member 25
DNAJC25-GNG10 readthrough
|
|
chr4_+_48038095
|
2.150
|
|
SLAIN2
|
SLAIN motif family, member 2
|
|
chr2_+_187059277
|
2.132
|
|
ZC3H15
|
zinc finger CCCH-type containing 15
|
|
chr2_+_54195913
|
2.112
|
NM_138448
|
ACYP2
|
acylphosphatase 2, muscle type
|
|
chr6_+_7535404
|
2.085
|
NM_152551
|
SNRNP48
|
small nuclear ribonucleoprotein 48kDa (U11/U12)
|
|
chr2_+_232534535
|
2.073
|
NM_152383
|
DIS3L2
|
DIS3 mitotic control homolog (S. cerevisiae)-like 2
|
|
chr18_+_75967902
|
2.059
|
NM_014913
|
ADNP2
|
ADNP homeobox 2
|
|
chr11_+_12652427
|
2.028
|
NM_021961
|
TEAD1
|
TEA domain family member 1 (SV40 transcriptional enhancer factor)
|
|
chr16_+_55523248
|
2.026
|
NM_001010989
NM_001010990
NM_014685
|
HERPUD1
|
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1
|
|
chr2_+_187059237
|
1.940
|
|
ZC3H15
|
zinc finger CCCH-type containing 15
|
|
chr4_+_95592067
|
1.896
|
|
PDLIM5
|
PDZ and LIM domain 5
|
|
chr5_+_72830029
|
1.865
|
|
BTF3
|
basic transcription factor 3
|
|
chr5_+_72830003
|
1.778
|
NM_001037637
NM_001207
|
BTF3
|
basic transcription factor 3
|
|
chr2_+_187059115
|
1.732
|
NM_018471
|
ZC3H15
|
zinc finger CCCH-type containing 15
|
|
chr12_+_68923477
|
1.705
|
|
CNOT2
|
CCR4-NOT transcription complex, subunit 2
|
|
chr14_+_31616196
|
1.671
|
NM_001030055
NM_001173
|
ARHGAP5
|
Rho GTPase activating protein 5
|
|
chr10_+_103995297
|
1.618
|
NM_004193
|
GBF1
|
golgi brefeldin A resistant guanine nucleotide exchange factor 1
|
|
chr10_+_103995248
|
1.609
|
|
GBF1
|
golgi brefeldin A resistant guanine nucleotide exchange factor 1
|
|
chr4_+_24844749
|
1.606
|
NM_018323
|
PI4K2B
|
phosphatidylinositol 4-kinase type 2 beta
|
|
chr2_+_233270275
|
1.603
|
|
GIGYF2
|
GRB10 interacting GYF protein 2
|
|
chr2_+_233270241
|
1.585
|
NM_001103146
NM_001103147
NM_001103148
NM_015575
|
GIGYF2
|
GRB10 interacting GYF protein 2
|
|
chr2_+_99320181
|
1.581
|
NM_015904
|
EIF5B
|
eukaryotic translation initiation factor 5B
|
|
chr12_+_119418215
|
1.574
|
NM_001037495
NM_003746
|
DYNLL1
|
dynein, light chain, LC8-type 1
|
|
chr12_+_119418299
|
1.573
|
|
DYNLL1
|
dynein, light chain, LC8-type 1
|
|
chr5_+_36912656
|
1.572
|
|
NIPBL
|
Nipped-B homolog (Drosophila)
|
|
chr10_+_103103797
|
1.571
|
NM_003939
NM_033637
|
BTRC
|
beta-transducin repeat containing
|
|
chr4_+_110574420
|
1.569
|
|
SEC24B
|
SEC24 family, member B (S. cerevisiae)
|
|
chr5_+_93980108
|
1.562
|
NM_032290
|
ANKRD32
|
ankyrin repeat domain 32
|
|
chr1_+_78243181
|
1.557
|
NM_007034
|
DNAJB4
|
DnaJ (Hsp40) homolog, subfamily B, member 4
|
|
chr16_+_19032744
|
1.553
|
NM_001034841
|
ITPRIPL2
|
inositol 1,4,5-triphosphate receptor interacting protein-like 2
|
|
chr18_+_9126742
|
1.529
|
NM_001083625
NM_015208
|
ANKRD12
|
ankyrin repeat domain 12
|
|
chr15_+_76517623
|
1.523
|
|
IREB2
|
iron-responsive element binding protein 2
|
|
chr14_+_51525942
|
1.500
|
NM_016039
|
C14orf166
|
chromosome 14 open reading frame 166
|
|
chr14_+_51526071
|
1.489
|
|
C14orf166
|
chromosome 14 open reading frame 166
|
|
chr5_+_65475760
|
1.481
|
NM_001077199
|
SREK1
|
splicing regulatory glutamine/lysine-rich protein 1
|
|
chr10_+_49184658
|
1.476
|
|
MAPK8
|
mitogen-activated protein kinase 8
|
|
chr13_+_26029879
|
1.470
|
|
WASF3
|
WAS protein family, member 3
|
|
chr3_+_151013672
|
1.461
|
|
RNF13
|
ring finger protein 13
|
|
chr12_+_119418294
|
1.452
|
|
DYNLL1
|
dynein, light chain, LC8-type 1
|
|
chr4_+_38545741
|
1.451
|
NM_138389
|
FAM114A1
|
family with sequence similarity 114, member A1
|
|
chr10_+_112669349
|
1.444
|
|
SHOC2
|
soc-2 suppressor of clear homolog (C. elegans)
|
|
chr13_+_26029799
|
1.439
|
NM_006646
|
WASF3
|
WAS protein family, member 3
|
|
chr7_+_130663134
|
1.436
|
NM_013255
|
MKLN1
|
muskelin 1, intracellular mediator containing kelch motifs
|
|
chr13_+_48448950
|
1.435
|
|
FNDC3A
|
fibronectin type III domain containing 3A
|
|
chr4_+_48038259
|
1.435
|
NM_020846
|
SLAIN2
|
SLAIN motif family, member 2
|
|
chr10_-_69762654
|
1.428
|
NM_001033083
NM_022129
|
PBLD
|
phenazine biosynthesis-like protein domain containing
|
|
chr4_+_110574376
|
1.423
|
NM_001042734
NM_006323
|
SEC24B
|
SEC24 family, member B (S. cerevisiae)
|
|
chr5_+_65475891
|
1.414
|
|
SREK1
|
splicing regulatory glutamine/lysine-rich protein 1
|
|
chr2_+_113119985
|
1.399
|
NM_005415
|
SLC20A1
|
solute carrier family 20 (phosphate transporter), member 1
|
|
chr4_-_110574363
|
1.378
|
|
1/2-SBSRNA4
|
lncRNA_BC009800
|
|
chr3_+_151013691
|
1.368
|
|
RNF13
|
ring finger protein 13
|
|
chr11_+_82290384
|
1.366
|
NM_145018
|
C11orf82
|
chromosome 11 open reading frame 82
|
|
chr10_+_112669353
|
1.356
|
|
SHOC2
|
soc-2 suppressor of clear homolog (C. elegans)
|
|
chr15_-_47235057
|
1.349
|
NM_001143887
NM_004236
|
COPS2
|
COP9 constitutive photomorphogenic homolog subunit 2 (Arabidopsis)
|
|
chr20_+_56660212
|
1.348
|
|
STX16
|
syntaxin 16
|
|
chr1_+_170768914
|
1.331
|
NM_016227
|
C1orf9
|
chromosome 1 open reading frame 9
|
|
chr4_+_128870968
|
1.303
|
NM_031291
|
SLC25A31
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 31
|
|
chr4_+_110574274
|
1.282
|
|
SEC24B
|
SEC24 family, member B (S. cerevisiae)
|
|
chr6_+_53038181
|
1.245
|
NM_033480
|
FBXO9
|
F-box protein 9
|
|
chr4_+_95592030
|
1.237
|
NM_001011513
NM_001011515
NM_001011516
NM_006457
|
PDLIM5
|
PDZ and LIM domain 5
|
|
chr1_+_170768635
|
1.236
|
NM_014283
|
C1orf9
|
chromosome 1 open reading frame 9
|
|
chr11_-_27341251
|
1.228
|
NM_030771
NM_080654
|
CCDC34
|
coiled-coil domain containing 34
|
|
chr10_+_93673485
|
1.225
|
|
BTAF1
|
BTAF1 RNA polymerase II, B-TFIID transcription factor-associated, 170kDa (Mot1 homolog, S. cerevisiae)
|
|
chr1_+_40399642
|
1.220
|
|
RLF
|
rearranged L-myc fusion
|
|
chr5_+_36912595
|
1.215
|
NM_015384
NM_133433
|
NIPBL
|
Nipped-B homolog (Drosophila)
|
|
chr13_+_48448912
|
1.205
|
|
FNDC3A
|
fibronectin type III domain containing 3A
|
|
chr3_+_49952480
|
1.204
|
NM_001167582
NM_005777
|
RBM6
|
RNA binding motif protein 6
|
|
chr2_+_233270286
|
1.201
|
|
GIGYF2
|
GRB10 interacting GYF protein 2
|
|
chr1_+_40399618
|
1.201
|
NM_012421
|
RLF
|
rearranged L-myc fusion
|
|
chr17_+_25730086
|
1.196
|
|
CPD
|
carboxypeptidase D
|
|
chr17_+_40581704
|
1.190
|
|
HEXIM1
|
hexamethylene bis-acetamide inducible 1
|
|
chr8_-_17148637
|
1.177
|
NM_013354
NM_054026
|
CNOT7
|
CCR4-NOT transcription complex, subunit 7
|
|
chr16_-_56038771
|
1.175
|
NM_020313
|
CIAPIN1
|
cytokine induced apoptosis inhibitor 1
|
|
chr16_-_55016803
|
1.174
|
|
AMFR
|
autocrine motility factor receptor
|
|
chr17_+_25730048
|
1.163
|
|
CPD
|
carboxypeptidase D
|
|
chr2_+_36436316
|
1.152
|
|
CRIM1
|
cysteine rich transmembrane BMP regulator 1 (chordin-like)
|
|
chr5_+_99899133
|
1.143
|
|
FAM174A
|
family with sequence similarity 174, member A
|
|
chr2_-_68547893
|
1.140
|
NM_001024680
|
FBXO48
|
F-box protein 48
|
|
chr8_-_38245694
|
1.138
|
NM_001102559
NM_001102560
NM_032483
|
PPAPDC1B
|
phosphatidic acid phosphatase type 2 domain containing 1B
|
|
chr14_-_91372365
|
1.134
|
NM_001128595
NM_152332
|
TC2N
|
tandem C2 domains, nuclear
|
|
chr2_+_46623325
|
1.128
|
NM_012249
|
RHOQ
|
ras homolog gene family, member Q
|
|
chr8_+_9450849
|
1.123
|
NM_003747
|
TNKS
|
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase
|
|
chr6_+_148705646
|
1.120
|
|
SASH1
|
SAM and SH3 domain containing 1
|
|
chr18_+_75968133
|
1.108
|
|
ADNP2
|
ADNP homeobox 2
|
|
chr10_+_93548119
|
1.098
|
NM_025235
|
TNKS2
|
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase 2
|
|
chr6_+_99389300
|
1.097
|
NM_005604
|
POU3F2
|
POU class 3 homeobox 2
|
|
chr10_+_93548261
|
1.096
|
|
TNKS2
|
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase 2
|
|
chr10_+_93548048
|
1.093
|
|
TNKS2
|
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase 2
|
|
chr5_+_179180447
|
1.081
|
NM_003900
|
SQSTM1
|
sequestosome 1
|
|
chr10_+_93548162
|
1.073
|
|
TNKS2
|
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase 2
|
|
chr2_+_27848090
|
1.070
|
|
MRPL33
|
mitochondrial ribosomal protein L33
|
|
chr1_-_183210155
|
1.063
|
|
FAM129A
|
family with sequence similarity 129, member A
|
|
chr20_-_48980858
|
1.062
|
NM_015339
NM_181442
|
ADNP
|
activity-dependent neuroprotector homeobox
|
|
chr22_+_28806452
|
1.055
|
NM_152510
|
HORMAD2
|
HORMA domain containing 2
|
|
chr2_+_190014403
|
1.047
|
NM_032168
|
WDR75
|
WD repeat domain 75
|
|
chr13_+_20770248
|
1.042
|
|
LOC650794
|
hypothetical LOC650794
|
|
chr12_+_87060176
|
1.033
|
NM_181783
|
TMTC3
|
transmembrane and tetratricopeptide repeat containing 3
|
|
chr10_+_112669267
|
1.032
|
NM_007373
|
SHOC2
|
soc-2 suppressor of clear homolog (C. elegans)
|
|
chr18_+_17076166
|
1.031
|
NM_001142966
|
GREB1L
|
growth regulation by estrogen in breast cancer-like
|
|
chr22_+_39817733
|
1.028
|
|
EP300
|
E1A binding protein p300
|
|
chr16_+_56038837
|
1.023
|
NM_020312
|
COQ9
|
coenzyme Q9 homolog (S. cerevisiae)
|
|
chr17_+_25730107
|
1.013
|
NM_001304
|
CPD
|
carboxypeptidase D
|
|
chr4_+_148757958
|
1.005
|
NM_018241
|
TMEM184C
|
transmembrane protein 184C
|
|
chr14_+_49157150
|
0.994
|
NM_002408
|
MGAT2
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase
|
|
chr3_+_184648089
|
0.994
|
|
LOC100505687
|
hypothetical LOC100505687
|
|
chr9_+_91123204
|
0.993
|
NM_024077
|
SECISBP2
|
SECIS binding protein 2
|
|
chr17_-_35273947
|
0.993
|
NM_012481
NM_183228
NM_183229
NM_183230
NM_183231
NM_183232
|
IKZF3
|
IKAROS family zinc finger 3 (Aiolos)
|
|
chr2_+_54536780
|
0.992
|
NM_003128
|
SPTBN1
|
spectrin, beta, non-erythrocytic 1
|
|
chr14_+_30564433
|
0.990
|
NM_001128126
NM_007077
|
AP4S1
|
adaptor-related protein complex 4, sigma 1 subunit
|
|
chr2_+_27848085
|
0.990
|
NM_004891
NM_145330
|
MRPL33
|
mitochondrial ribosomal protein L33
|
|
chr6_+_53038176
|
0.982
|
|
FBXO9
|
F-box protein 9
|
|
chr2_+_46623396
|
0.982
|
|
RHOQ
|
ras homolog gene family, member Q
|
|
chr15_+_47235243
|
0.975
|
NM_001001556
|
GALK2
|
galactokinase 2
|
|
chr6_+_53038119
|
0.964
|
|
FBXO9
|
F-box protein 9
|
|
chr5_-_36912487
|
0.958
|
|
LOC646719
|
hypothetical LOC646719
|
|
chr17_+_43081837
|
0.949
|
|
KPNB1
|
karyopherin (importin) beta 1
|
|
chr3_-_27385856
|
0.945
|
NM_199347
|
NEK10
|
NIMA (never in mitosis gene a)- related kinase 10
|
|
chr1_+_7753913
|
0.941
|
NM_004781
|
VAMP3
|
vesicle-associated membrane protein 3 (cellubrevin)
|
|
chr2_+_190014432
|
0.940
|
|
WDR75
|
WD repeat domain 75
|
|
chr5_+_112340326
|
0.931
|
NM_152624
|
DCP2
|
DCP2 decapping enzyme homolog (S. cerevisiae)
|
|
chr8_+_26204921
|
0.930
|
NM_002717
|
PPP2R2A
|
protein phosphatase 2, regulatory subunit B, alpha
|
|
chr1_+_74436507
|
0.926
|
NM_001112808
NM_003838
|
FPGT-TNNI3K
FPGT
|
FPGT-TNNI3K fusion protein
fucose-1-phosphate guanylyltransferase
|
|
chr20_+_57948803
|
0.923
|
NM_001190827
NM_022106
|
C20orf177
|
chromosome 20 open reading frame 177
|
|
chr12_+_8741746
|
0.918
|
NM_020734
|
RIMKLB
|
ribosomal modification protein rimK-like family member B
|
|
chr11_+_82460766
|
0.915
|
|
LOC100506233
|
hypothetical LOC100506233
|
|
chr2_-_198072868
|
0.911
|
NM_002156
|
HSPD1
|
heat shock 60kDa protein 1 (chaperonin)
|
|
chr9_-_32991518
|
0.909
|
NM_001195249
NM_001195254
NM_001195248
NM_001195250
NM_001195251
NM_001195252
NM_175069
NM_175073
|
APTX
|
aprataxin
|
|
chr2_+_152900076
|
0.908
|
|
FMNL2
|
formin-like 2
|
|
chr12_+_32151351
|
0.908
|
NM_001003398
NM_001714
|
BICD1
|
bicaudal D homolog 1 (Drosophila)
|
|
chr3_+_138063764
|
0.905
|
|
NCK1
|
NCK adaptor protein 1
|
|
chr7_+_33135376
|
0.903
|
NM_001033604
NM_001033605
NM_014451
NM_198428
|
BBS9
|
Bardet-Biedl syndrome 9
|
|
chr5_+_179180509
|
0.903
|
|
SQSTM1
|
sequestosome 1
|
|
chr3_+_143925904
|
0.903
|
NM_003304
|
TRPC1
|
transient receptor potential cation channel, subfamily C, member 1
|
|
chr6_+_99389318
|
0.901
|
|
POU3F2
|
POU class 3 homeobox 2
|
|
chr2_+_68548173
|
0.901
|
NM_173545
|
APLF
|
aprataxin and PNKP like factor
|
|
chr4_-_122521562
|
0.896
|
NM_198179
|
QRFPR
|
pyroglutamylated RFamide peptide receptor
|
|
chr4_+_148758014
|
0.892
|
|
TMEM184C
|
transmembrane protein 184C
|
|
chr16_-_55016872
|
0.887
|
NM_001144
|
AMFR
|
autocrine motility factor receptor
|
|
chr8_+_59628372
|
0.883
|
|
SDCBP
|
syndecan binding protein (syntenin)
|
|
chr11_+_125279481
|
0.883
|
NM_013264
|
DDX25
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 25
|
|
chr6_+_117305068
|
0.882
|
NM_173560
|
RFX6
|
regulatory factor X, 6
|
|
chr8_+_59628352
|
0.881
|
|
SDCBP
|
syndecan binding protein (syntenin)
|
|
chr4_+_148621567
|
0.879
|
|
EDNRA
|
endothelin receptor type A
|
|
chr4_+_81475897
|
0.874
|
NM_152770
|
C4orf22
|
chromosome 4 open reading frame 22
|
|
chr5_+_80292213
|
0.872
|
|
RASGRF2
|
Ras protein-specific guanine nucleotide-releasing factor 2
|
|
chr8_+_9450832
|
0.872
|
|
TNKS
|
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase
|
|
chr3_-_99795074
|
0.868
|
NM_000097
|
CPOX
|
coproporphyrinogen oxidase
|
|
chr1_+_181708235
|
0.866
|
|
SMG7
|
Smg-7 homolog, nonsense mediated mRNA decay factor (C. elegans)
|
|
chr10_+_93673544
|
0.866
|
NM_003972
|
BTAF1
|
BTAF1 RNA polymerase II, B-TFIID transcription factor-associated, 170kDa (Mot1 homolog, S. cerevisiae)
|
|
chr2_+_223433919
|
0.862
|
NM_004457
NM_203372
|
ACSL3
|
acyl-CoA synthetase long-chain family member 3
|
|
chr13_+_48448628
|
0.858
|
|
FNDC3A
|
fibronectin type III domain containing 3A
|
|
chr5_+_179180517
|
0.856
|
|
SQSTM1
|
sequestosome 1
|
|
chr1_+_52380633
|
0.849
|
NM_004799
NM_007323
NM_007324
|
ZFYVE9
|
zinc finger, FYVE domain containing 9
|
|
chr16_-_69822058
|
0.843
|
NM_032821
NM_017558
|
HYDIN
|
hydrocephalus inducing homolog (mouse)
|
|
chr7_-_84654106
|
0.843
|
|
SEMA3D
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3D
|
|
chr5_+_99898922
|
0.842
|
NM_198507
|
FAM174A
|
family with sequence similarity 174, member A
|
|
chr17_-_46553083
|
0.841
|
NM_001130528
NM_003971
|
SPAG9
|
sperm associated antigen 9
|
|
chr4_+_164635291
|
0.840
|
|
C4orf43
|
chromosome 4 open reading frame 43
|
|
chr3_-_181237210
|
0.839
|
NM_016559
|
PEX5L
|
peroxisomal biogenesis factor 5-like
|
|
chrX_+_105856531
|
0.838
|
NM_194463
|
RNF128
|
ring finger protein 128
|
|
chr2_-_202216451
|
0.837
|
NM_001044385
|
ALS2CR4
|
amyotrophic lateral sclerosis 2 (juvenile) chromosome region, candidate 4
|
|
chr4_+_164635263
|
0.836
|
|
C4orf43
|
chromosome 4 open reading frame 43
|
|
chr10_+_127575097
|
0.834
|
NM_145235
|
FANK1
|
fibronectin type III and ankyrin repeat domains 1
|
|
chr8_+_17148755
|
0.830
|
NM_001145152
NM_152415
|
VPS37A
|
vacuolar protein sorting 37 homolog A (S. cerevisiae)
|
|
chr1_+_24842180
|
0.829
|
NM_005839
|
SRRM1
|
serine/arginine repetitive matrix 1
|
|
chr12_-_6586736
|
0.829
|
|
CHD4
|
chromodomain helicase DNA binding protein 4
|
|
chr14_-_54563554
|
0.829
|
NM_001008396
NM_007086
|
WDHD1
|
WD repeat and HMG-box DNA binding protein 1
|
|
chr3_+_98966253
|
0.828
|
NM_032146
NM_177976
|
ARL6
|
ADP-ribosylation factor-like 6
|
|
chr13_+_30089829
|
0.825
|
NM_005800
|
USPL1
|
ubiquitin specific peptidase like 1
|
|
chr4_-_86106567
|
0.824
|
NM_014991
|
WDFY3
|
WD repeat and FYVE domain containing 3
|
|
chr15_+_63948823
|
0.823
|
NM_004663
|
RAB11A
|
RAB11A, member RAS oncogene family
|
|
chr14_+_74030195
|
0.821
|
NM_194279
|
ISCA2
|
iron-sulfur cluster assembly 2 homolog (S. cerevisiae)
|
|
chr10_-_127574957
|
0.821
|
|
DHX32
|
DEAH (Asp-Glu-Ala-His) box polypeptide 32
|
|
chr7_+_30034731
|
0.811
|
|
PLEKHA8P1
PLEKHA8
|
pleckstrin homology domain containing, family A member 8 pseudogene 1
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 8
|
|
chr5_+_131920455
|
0.811
|
NM_005732
|
RAD50
|
RAD50 homolog (S. cerevisiae)
|
|
chr5_+_156819782
|
0.811
|
|
NIPAL4
|
NIPA-like domain containing 4
|
|
chr4_+_164635298
|
0.811
|
|
C4orf43
|
chromosome 4 open reading frame 43
|
|
chr10_+_98582005
|
0.806
|
NM_001170765
NM_001170766
|
LCOR
|
ligand dependent nuclear receptor corepressor
|
|
chr12_+_122635052
|
0.805
|
|
TMED2
|
transmembrane emp24 domain trafficking protein 2
|
|
chr8_+_59628281
|
0.805
|
NM_001007067
NM_001007068
NM_001007069
NM_001007070
NM_005625
|
SDCBP
|
syndecan binding protein (syntenin)
|
|
chr15_+_73923047
|
0.801
|
|
UBE2Q2
|
ubiquitin-conjugating enzyme E2Q family member 2
|
|
chr3_+_12573513
|
0.801
|
NM_014160
|
MKRN2
|
makorin ring finger protein 2
|
|
chr8_+_9450800
|
0.799
|
|
TNKS
|
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase
|
|
chr1_+_24842298
|
0.799
|
|
SRRM1
|
serine/arginine repetitive matrix 1
|
|
chr1_+_176329895
|
0.796
|
|
RASAL2
|
RAS protein activator like 2
|
|
chr5_-_99898788
|
0.796
|
|
|
|
|
chr4_+_164635270
|
0.791
|
|
C4orf43
|
chromosome 4 open reading frame 43
|
|
chr10_-_69661860
|
0.787
|
NM_145178
|
ATOH7
|
atonal homolog 7 (Drosophila)
|
|
chr3_-_109423858
|
0.787
|
NM_018010
|
IFT57
|
intraflagellar transport 57 homolog (Chlamydomonas)
|
|
chr9_+_33015287
|
0.779
|
|
DNAJA1
|
DnaJ (Hsp40) homolog, subfamily A, member 1
|
|
chr8_+_59628337
|
0.779
|
|
SDCBP
|
syndecan binding protein (syntenin)
|
|
chr8_-_110415448
|
0.778
|
NM_032869
|
NUDCD1
|
NudC domain containing 1
|
|
chr7_+_23111928
|
0.772
|
|
KLHL7
|
kelch-like 7 (Drosophila)
|
|
chr6_+_7052798
|
0.769
|
NM_001168344
|
RREB1
|
ras responsive element binding protein 1
|