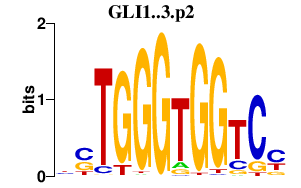
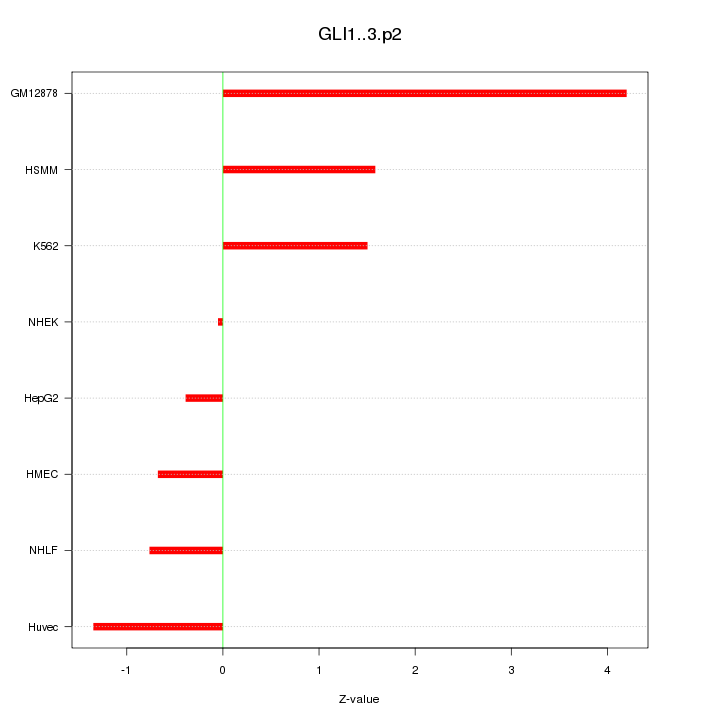
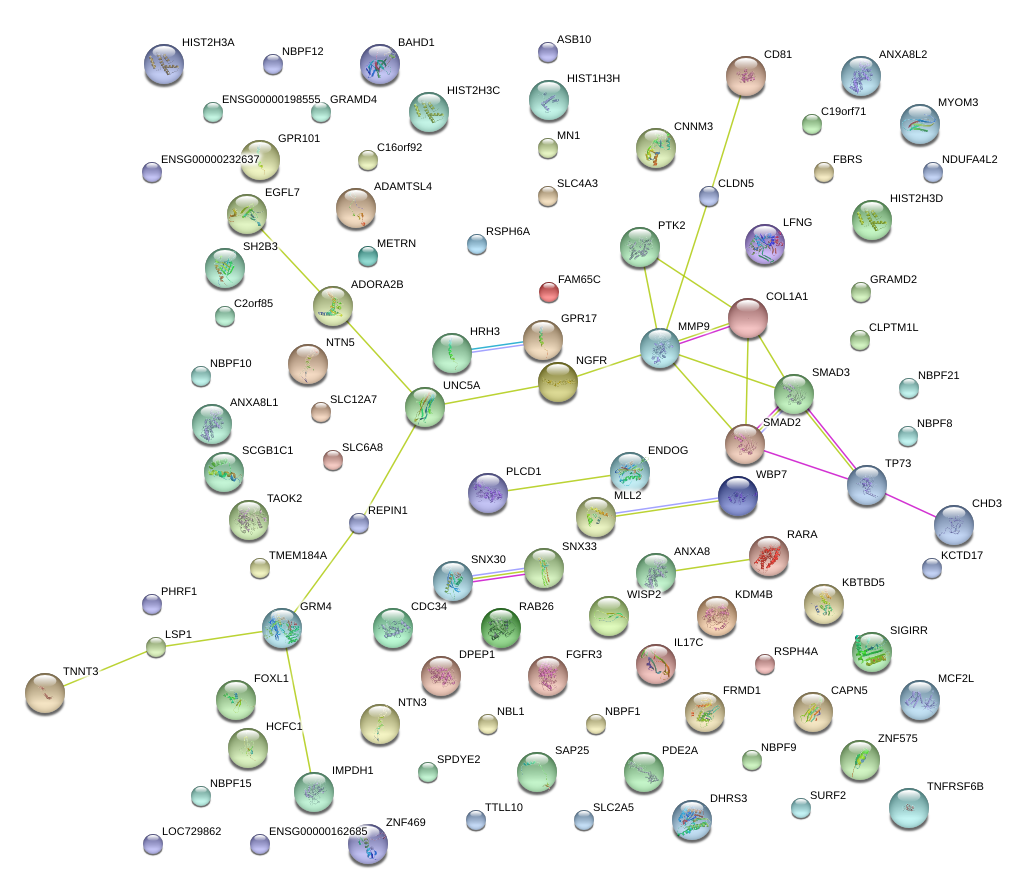
|
chr11_+_566482
|
6.628
|
NM_020901
|
PHRF1
|
PHD and ring finger domains 1
|
|
chr16_+_2461500
|
2.748
|
NM_006181
|
NTN3
|
netrin 3
|
|
chr7_-_100009205
|
2.537
|
NM_001168682
|
SAP25
|
sin3A-binding protein 25
|
|
chr15_+_65217412
|
2.504
|
NM_001145103
|
SMAD3
|
SMAD family member 3
|
|
chr4_+_1764802
|
2.456
|
NM_000142
NM_001163213
NM_022965
|
FGFR3
|
fibroblast growth factor receptor 3
|
|
chr1_+_148090804
|
2.323
|
NM_021059
NM_001005464
|
HIST2H3C
HIST2H3A
|
histone cluster 2, H3c
histone cluster 2, H3a
|
|
chr11_-_72057755
|
2.297
|
NM_001146209
|
PDE2A
|
phosphodiesterase 2A, cGMP-stimulated
|
|
chr11_+_1848674
|
2.216
|
NM_001013254
NM_001013255
|
LSP1
|
lymphocyte-specific protein 1
|
|
chr22_+_18090118
|
2.067
|
|
SEPT5-GP1BB
|
SEPT5-GP1BB read-through transcript
|
|
chr13_+_112789669
|
2.040
|
|
MCF2L
|
MCF.2 cell line derived transforming sequence-like
|
|
chr22_-_17892001
|
2.033
|
|
CLDN5
|
claudin 5
|
|
chr12_+_110328126
|
2.029
|
NM_005475
|
SH2B3
|
SH2B adaptor protein 3
|
|
chr11_+_76491532
|
1.946
|
NM_006189
|
OMP
|
olfactory marker protein
|
|
chr19_-_51010272
|
1.936
|
NM_030785
|
RSPH6A
|
radial spoke head 6 homolog A (Chlamydomonas)
|
|
chr11_-_404975
|
1.935
|
NM_001135054
|
SIGIRR
|
single immunoglobulin and toll-interleukin 1 receptor (TIR) domain
|
|
chr16_+_2138608
|
1.898
|
NM_014353
|
RAB26
|
RAB26, member RAS oncogene family
|
|
chr6_-_34209250
|
1.869
|
NM_000841
|
GRM4
|
glutamate receptor, metabotropic 4
|
|
chr5_+_176170041
|
1.803
|
NM_133369
|
UNC5A
|
unc-5 homolog A (C. elegans)
|
|
chr19_+_482737
|
1.803
|
|
CDC34
|
cell division cycle 34 homolog (S. cerevisiae)
|
|
chr19_+_3490154
|
1.794
|
NM_001135580
|
C19orf71
|
chromosome 19 open reading frame 71
|
|
chr16_+_87232501
|
1.761
|
NM_013278
|
IL17C
|
interleukin 17C
|
|
chr11_+_1897374
|
1.755
|
NM_001042780
NM_001042781
NM_001042782
NM_006757
|
TNNT3
|
troponin T type 3 (skeletal, fast)
|
|
chr7_+_149699185
|
1.754
|
NM_014374
|
REPIN1
|
replication initiator 1
|
|
chr7_-_150515851
|
1.749
|
NM_080871
|
ASB10
|
ankyrin repeat and SOCS box containing 10
|
|
chr1_-_12599934
|
1.745
|
NM_004753
|
DHRS3
|
dehydrogenase/reductase (SDR family) member 3
|
|
chr22_-_17891777
|
1.705
|
|
CLDN5
|
claudin 5
|
|
chr22_-_26527469
|
1.692
|
NM_002430
|
MN1
|
meningioma (disrupted in balanced translocation) 1
|
|
chr2_+_128119908
|
1.679
|
NM_001161415
NM_001161416
NM_001161417
NM_005291
|
GPR17
|
G protein-coupled receptor 17
|
|
chr5_-_1165106
|
1.665
|
NM_006598
|
SLC12A7
|
solute carrier family 12 (potassium/chloride transporters), member 7
|
|
chr16_+_88214500
|
1.623
|
NM_004413
|
DPEP1
|
dipeptidase 1 (renal)
|
|
chr17_+_15788955
|
1.598
|
NM_000676
|
ADORA2B
|
adenosine A2b receptor
|
|
chr3_-_38046034
|
1.587
|
NM_006225
|
PLCD1
|
phospholipase C, delta 1
|
|
chr7_+_101978777
|
1.572
|
NM_001166339
NM_001031618
|
SPDYE2L
SPDYE2
|
WBSCR19-like protein 3
speedy homolog E2 (Xenopus laevis)
|
|
chr11_+_183079
|
1.563
|
NM_145651
|
SCGB1C1
|
secretoglobin, family 1C, member 1
|
|
chr8_-_142080169
|
1.557
|
NM_005607
NM_153831
|
PTK2
|
PTK2 protein tyrosine kinase 2
|
|
chr1_-_24307514
|
1.543
|
|
MYOM3
|
myomesin family, member 3
|
|
chr2_+_220200530
|
1.509
|
NM_005070
NM_201574
|
SLC4A3
|
solute carrier family 4, anion exchanger, member 3
|
|
chrX_-_135941498
|
1.509
|
NM_054021
|
GPR101
|
G protein-coupled receptor 101
|
|
chr22_+_45401321
|
1.506
|
NM_015124
|
GRAMD4
|
GRAM domain containing 4
|
|
chr20_-_48686832
|
1.503
|
NM_080829
|
FAM65C
|
family with sequence similarity 65, member C
|
|
chr17_+_35751796
|
1.500
|
NM_001024809
|
RARA
|
retinoic acid receptor, alpha
|
|
chr1_-_16812274
|
1.499
|
NM_017940
|
NBPF1
|
neuroblastoma breakpoint family, member 1
|
|
chr1_+_3597092
|
1.492
|
NM_001126240
NM_001126241
NM_001126242
|
TP73
|
tumor protein p73
|
|
chr17_+_44927591
|
1.492
|
NM_002507
|
NGFR
|
nerve growth factor receptor
|
|
chr2_+_242460553
|
1.482
|
NM_173821
|
C2orf85
|
chromosome 2 open reading frame 85
|
|
chr3_+_42702014
|
1.465
|
NM_152393
|
KBTBD5
|
kelch repeat and BTB (POZ) domain containing 5
|
|
chr9_+_130620540
|
1.464
|
NM_004435
|
ENDOG
|
endonuclease G
|
|
chr2_+_96845704
|
1.457
|
NM_017623
NM_199078
|
CNNM3
|
cyclin M3
|
|
chr1_-_9040141
|
1.421
|
|
SLC2A5
|
solute carrier family 2 (facilitated glucose/fructose transporter), member 5
|
|
chr6_-_168222645
|
1.396
|
NM_024919
|
FRMD1
|
FERM domain containing 1
|
|
chr7_-_1562272
|
1.381
|
NM_001097620
|
TMEM184A
|
transmembrane protein 184A
|
|
chr7_-_127833231
|
1.371
|
NM_001142573
NM_001142574
NM_001142575
|
IMPDH1
|
IMP (inosine 5'-monophosphate) dehydrogenase 1
|
|
chr20_+_42776888
|
1.370
|
|
WISP2
|
WNT1 inducible signaling pathway protein 2
|
|
chr17_+_7728819
|
1.370
|
NM_001005271
|
CHD3
|
chromodomain helicase DNA binding protein 3
|
|
chr11_+_76455574
|
1.370
|
NM_004055
|
CAPN5
|
calpain 5
|
|
chr16_+_29892714
|
1.364
|
NM_004783
NM_016151
|
TAOK2
|
TAO kinase 2
|
|
chr22_+_35777561
|
1.359
|
NM_024681
|
KCTD17
|
potassium channel tetramerisation domain containing 17
|
|
chr17_-_40674883
|
1.356
|
|
|
|
|
chr20_+_61798446
|
1.354
|
NM_003823
|
TNFRSF6B
|
tumor necrosis factor receptor superfamily, member 6b, decoy
|
|
chr19_+_48729127
|
1.354
|
NM_174945
|
ZNF575
|
zinc finger protein 575
|
|
chr20_+_42777298
|
1.352
|
NM_003881
|
WISP2
|
WNT1 inducible signaling pathway protein 2
|
|
chr7_+_2525922
|
1.345
|
NM_001040167
NM_001040168
|
LFNG
|
LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase
|
|
chr1_+_19842309
|
1.335
|
NM_182744
|
NBL1
|
neuroblastoma, suppression of tumorigenicity 1
|
|
chr20_+_61798512
|
1.330
|
|
TNFRSF6B
|
tumor necrosis factor receptor superfamily, member 6b, decoy
|
|
chr12_-_55920741
|
1.326
|
NM_020142
|
NDUFA4L2
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4-like 2
|
|
chr15_-_70277163
|
1.314
|
NM_001012642
|
GRAMD2
|
GRAM domain containing 2
|
|
chr15_+_38520594
|
1.297
|
NM_014952
|
BAHD1
|
bromo adjacent homology domain containing 1
|
|
chrX_-_152871351
|
1.296
|
|
HCFC1
|
host cell factor C1 (VP16-accessory protein)
|
|
chr17_+_73738985
|
1.290
|
NM_001145529
|
TMEM235
|
transmembrane protein 235
|
|
chr9_+_138677197
|
1.287
|
NM_201446
NM_016215
|
EGFL7
|
EGF-like-domain, multiple 7
|
|
chr1_+_1104939
|
1.283
|
NM_153254
|
TTLL10
|
tubulin tyrosine ligase-like family, member 10
|
|
chr19_+_4920123
|
1.277
|
NM_015015
|
KDM4B
|
lysine (K)-specific demethylase 4B
|
|
chr17_-_45617899
|
1.276
|
|
COL1A1
|
collagen, type I, alpha 1
|
|
chr10_+_47216925
|
1.273
|
NM_001630
|
ANXA8L2
ANXA8
|
annexin A8-like 2
annexin A8
|
|
chr16_+_705080
|
1.267
|
NM_024042
|
METRN
|
meteorin, glial cell differentiation regulator
|
|
chr12_+_6517600
|
1.264
|
|
|
|
|
chr20_+_44070946
|
1.264
|
NM_004994
|
MMP9
|
matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase)
|
|
chr16_+_30583278
|
1.264
|
NM_001105079
|
FBRS
|
fibrosin
|
|
chr5_-_1397941
|
1.263
|
NM_030782
|
CLPTM1L
|
CLPTM1-like
|
|
chr9_+_114552871
|
1.261
|
NM_001012994
|
SNX30
|
sorting nexin family member 30
|
|
chr20_-_60228665
|
1.259
|
NM_007232
|
HRH3
|
histamine receptor H3
|
|
chr19_+_4920126
|
1.257
|
|
KDM4B
|
lysine (K)-specific demethylase 4B
|
|
chr1_+_148788410
|
1.244
|
NM_019032
NM_025008
|
ADAMTSL4
|
ADAMTS-like 4
|
|
chr16_+_87021379
|
1.218
|
NM_001127464
|
ZNF469
|
zinc finger protein 469
|
|
chr17_+_45617676
|
1.217
|
|
|
|
|
chr19_+_40900965
|
1.203
|
|
MLL4
|
myeloid/lymphoid or mixed-lineage leukemia 4
|
|
chr19_-_53868075
|
1.202
|
NM_145807
|
NTN5
|
netrin 5
|
|
chr16_+_85169615
|
1.202
|
NM_005250
|
FOXL1
|
forkhead box L1
|
|
chr9_+_135213235
|
1.201
|
NM_017503
|
SURF2
|
surfeit 2
|
|
chr16_+_29942155
|
1.199
|
NM_001109659
NM_001109660
|
C16orf92
|
chromosome 16 open reading frame 92
|
|
chrX_+_152608159
|
1.190
|
NM_001142806
|
SLC6A8
|
solute carrier family 6 (neurotransmitter transporter, creatine), member 8
|
|
chr11_+_2355119
|
1.178
|
NM_004356
|
CD81
|
CD81 molecule
|
|
chr10_+_80743769
|
1.175
|
|
ZMIZ1
|
zinc finger, MIZ-type containing 1
|
|
chr20_+_61655934
|
1.173
|
|
|
|
|
chr17_-_37022465
|
1.172
|
NM_005557
|
KRT16
|
keratin 16
|
|
chr19_+_40900757
|
1.166
|
NM_014727
|
MLL4
|
myeloid/lymphoid or mixed-lineage leukemia 4
|
|
chr20_+_60558104
|
1.143
|
NM_178463
|
C20orf166
|
chromosome 20 open reading frame 166
|
|
chr16_-_2258067
|
1.141
|
|
RNPS1
|
RNA binding protein S1, serine-rich domain
|
|
chr14_+_105012106
|
1.140
|
NM_001312
|
CRIP2
|
cysteine-rich protein 2
|
|
chr1_+_43837096
|
1.138
|
|
PTPRF
|
protein tyrosine phosphatase, receptor type, F
|
|
chr16_+_30583599
|
1.133
|
|
FBRS
|
fibrosin
|
|
chr4_-_140696740
|
1.133
|
NM_030648
|
SETD7
|
SET domain containing (lysine methyltransferase) 7
|
|
chr12_-_51514328
|
1.127
|
NM_175834
|
KRT79
|
keratin 79
|
|
chr16_+_82782223
|
1.122
|
NM_001145400
NM_139174
|
ADAD2
|
adenosine deaminase domain containing 2
|
|
chr22_-_49050847
|
1.122
|
NM_002751
|
MAPK11
|
mitogen-activated protein kinase 11
|
|
chr19_+_16296632
|
1.119
|
NM_016270
|
KLF2
|
Kruppel-like factor 2 (lung)
|
|
chr1_-_38285033
|
1.115
|
NM_002699
|
POU3F1
|
POU class 3 homeobox 1
|
|
chr1_-_150065176
|
1.112
|
NM_001001523
|
RORC
|
RAR-related orphan receptor C
|
|
chr6_+_44076281
|
1.109
|
NM_001171992
NM_153246
|
C6orf223
|
chromosome 6 open reading frame 223
|
|
chr6_+_41622267
|
1.109
|
|
FOXP4
|
forkhead box P4
|
|
chr1_+_101475149
|
1.108
|
|
S1PR1
|
sphingosine-1-phosphate receptor 1
|
|
chr17_+_71141108
|
1.104
|
NM_001162995
|
LOC643008
|
hypothetical protein LOC643008
|
|
chr1_+_200358594
|
1.103
|
NM_004767
|
GPR37L1
|
G protein-coupled receptor 37 like 1
|
|
chr22_+_18388656
|
1.100
|
|
C22orf25
|
chromosome 22 open reading frame 25
|
|
chr15_+_72397934
|
1.097
|
NM_182791
|
CCDC33
|
coiled-coil domain containing 33
|
|
chr11_-_3104413
|
1.086
|
|
OSBPL5
|
oxysterol binding protein-like 5
|
|
chr8_+_21956335
|
1.078
|
NM_003867
|
FGF17
|
fibroblast growth factor 17
|
|
chr12_-_6668880
|
1.075
|
NM_001039917
NM_001039919
NM_001039920
NM_001135734
|
ZNF384
|
zinc finger protein 384
|
|
chr16_-_73842884
|
1.070
|
NM_001170717
NM_014567
|
BCAR1
|
breast cancer anti-estrogen resistance 1
|
|
chr9_-_138757175
|
1.069
|
NM_001001712
|
LCN10
|
lipocalin 10
|
|
chr20_+_42777419
|
1.054
|
|
WISP2
|
WNT1 inducible signaling pathway protein 2
|
|
chr3_+_50272648
|
1.046
|
|
|
|
|
chr18_-_72221228
|
1.044
|
|
ZNF516
|
zinc finger protein 516
|
|
chrX_-_19598912
|
1.041
|
NM_001184960
|
SH3KBP1
|
SH3-domain kinase binding protein 1
|
|
chr9_+_130056513
|
1.040
|
|
|
|
|
chr20_+_44180355
|
1.036
|
|
CD40
|
CD40 molecule, TNF receptor superfamily member 5
|
|
chr19_+_1236766
|
1.033
|
NM_001405
|
EFNA2
|
ephrin-A2
|
|
chr2_+_232281470
|
1.028
|
NM_001099285
NM_002823
|
PTMA
|
prothymosin, alpha
|
|
chr20_+_61654816
|
1.026
|
NM_024059
|
C20orf195
|
chromosome 20 open reading frame 195
|
|
chr22_+_18388633
|
1.026
|
|
C22orf25
|
chromosome 22 open reading frame 25
|
|
chr20_-_42457717
|
1.023
|
|
|
|
|
chr16_+_29581783
|
1.023
|
NM_001030288
|
SPN
|
sialophorin
|
|
chr10_+_80740584
|
1.018
|
|
ZMIZ1
|
zinc finger, MIZ-type containing 1
|
|
chr11_+_113351006
|
1.006
|
NM_000869
NM_213621
|
HTR3A
|
5-hydroxytryptamine (serotonin) receptor 3A
|
|
chr9_+_130909926
|
1.004
|
|
|
|
|
chr17_+_24095135
|
1.001
|
NM_004295
|
TRAF4
|
TNF receptor-associated factor 4
|
|
chr19_+_811615
|
0.991
|
|
CFD
|
complement factor D (adipsin)
|
|
chr12_-_6668680
|
0.990
|
|
ZNF384
|
zinc finger protein 384
|
|
chr1_+_11674365
|
0.988
|
NM_198545
|
C1orf187
|
chromosome 1 open reading frame 187
|
|
chr19_+_627346
|
0.985
|
NM_005860
|
FSTL3
|
follistatin-like 3 (secreted glycoprotein)
|
|
chr6_-_39390073
|
0.983
|
NM_031460
NM_001135111
|
KCNK17
|
potassium channel, subfamily K, member 17
|
|
chr11_+_65754476
|
0.981
|
|
PACS1
|
phosphofurin acidic cluster sorting protein 1
|
|
chr20_+_44180296
|
0.981
|
NM_001250
NM_152854
|
CD40
|
CD40 molecule, TNF receptor superfamily member 5
|
|
chr11_+_65236261
|
0.975
|
|
KAT5
|
K(lysine) acetyltransferase 5
|
|
chrX_+_69581429
|
0.966
|
NM_021120
|
DLG3
|
discs, large homolog 3 (Drosophila)
|
|
chr17_-_24302597
|
0.965
|
NM_001033561
NM_020889
|
PHF12
|
PHD finger protein 12
|
|
chr8_-_37847095
|
0.962
|
|
RAB11FIP1
|
RAB11 family interacting protein 1 (class I)
|
|
chr19_+_935316
|
0.958
|
NM_024100
|
WDR18
|
WD repeat domain 18
|
|
chr11_-_118693021
|
0.955
|
NM_006500
|
MCAM
|
melanoma cell adhesion molecule
|
|
chr8_-_22070208
|
0.953
|
NM_139278
|
LGI3
|
leucine-rich repeat LGI family, member 3
|
|
chr2_+_232281491
|
0.951
|
|
PTMA
|
prothymosin, alpha
|
|
chr17_-_17340219
|
0.948
|
NM_016084
|
RASD1
|
RAS, dexamethasone-induced 1
|
|
chr20_+_56861430
|
0.940
|
NM_001077490
NM_080425
|
GNAS
|
GNAS complex locus
|
|
chr19_+_63670148
|
0.936
|
NM_014347
|
ZNF324
|
zinc finger protein 324
|
|
chr19_+_10900502
|
0.932
|
|
C19orf52
|
chromosome 19 open reading frame 52
|
|
chr17_-_7078446
|
0.926
|
NM_004422
|
DVL2
|
dishevelled, dsh homolog 2 (Drosophila)
|
|
chr11_+_66367458
|
0.918
|
NM_001032279
NM_005133
|
RCE1
|
RCE1 homolog, prenyl protein peptidase (S. cerevisiae)
|
|
chr12_-_6668787
|
0.918
|
NM_001039918
NM_133476
|
ZNF384
|
zinc finger protein 384
|
|
chr3_-_185578621
|
0.918
|
NM_000460
NM_001177597
NM_001177598
|
THPO
|
thrombopoietin
|
|
chr19_+_13124889
|
0.917
|
|
IER2
|
immediate early response 2
|
|
chr17_-_74231240
|
0.917
|
|
CYTH1
|
cytohesin 1
|
|
chr22_+_18388602
|
0.914
|
NM_152906
|
C22orf25
|
chromosome 22 open reading frame 25
|
|
chr17_+_77603064
|
0.911
|
|
GPS1
|
G protein pathway suppressor 1
|
|
chr11_-_66367279
|
0.909
|
|
|
|
|
chr10_+_80672842
|
0.909
|
|
ZMIZ1
|
zinc finger, MIZ-type containing 1
|
|
chr1_+_3378030
|
0.908
|
|
ARHGEF16
|
Rho guanine nucleotide exchange factor (GEF) 16
|
|
chr20_-_62208627
|
0.907
|
NM_005286
|
NPBWR2
|
neuropeptides B/W receptor 2
|
|
chr17_+_18069593
|
0.903
|
NM_004140
|
LLGL1
|
lethal giant larvae homolog 1 (Drosophila)
|
|
chr22_-_17546234
|
0.898
|
|
SLC25A1
|
solute carrier family 25 (mitochondrial carrier; citrate transporter), member 1
|
|
chrX_+_151837064
|
0.896
|
NM_001178113
|
ZNF185
|
zinc finger protein 185 (LIM domain)
|
|
chr14_+_99181232
|
0.894
|
NM_001127258
NM_032425
|
HHIPL1
|
HHIP-like 1
|
|
chr22_-_17546296
|
0.893
|
NM_005984
|
SLC25A1
|
solute carrier family 25 (mitochondrial carrier; citrate transporter), member 1
|
|
chr12_+_124115877
|
0.891
|
NM_023928
|
AACS
|
acetoacetyl-CoA synthetase
|
|
chr22_-_17546276
|
0.889
|
|
SLC25A1
|
solute carrier family 25 (mitochondrial carrier; citrate transporter), member 1
|
|
chr16_+_11669736
|
0.883
|
NM_003498
|
SNN
|
stannin
|
|
chr17_-_40401138
|
0.876
|
NM_006688
|
C1QL1
|
complement component 1, q subcomponent-like 1
|
|
chr19_-_3820012
|
0.876
|
NM_001145640
NM_015174
|
ZFR2
|
zinc finger RNA binding protein 2
|
|
chr16_-_82778071
|
0.874
|
NM_005679
NM_139353
|
TAF1C
|
TATA box binding protein (TBP)-associated factor, RNA polymerase I, C, 110kDa
|
|
chr1_+_32251988
|
0.872
|
NM_006559
|
KHDRBS1
|
KH domain containing, RNA binding, signal transduction associated 1
|
|
chr19_+_16296734
|
0.867
|
|
KLF2
|
Kruppel-like factor 2 (lung)
|
|
chr17_+_14145051
|
0.865
|
NM_006041
|
HS3ST3B1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 3B1
|
|
chr8_-_144726064
|
0.864
|
NM_001100878
|
C8orf73
|
chromosome 8 open reading frame 73
|
|
chr11_+_65815940
|
0.862
|
NM_153266
|
TMEM151A
|
transmembrane protein 151A
|
|
chr1_+_1256585
|
0.862
|
NM_152228
|
TAS1R3
|
taste receptor, type 1, member 3
|
|
chr19_-_10900086
|
0.861
|
NM_024029
|
YIPF2
|
Yip1 domain family, member 2
|
|
chr19_+_13124870
|
0.852
|
|
IER2
|
immediate early response 2
|
|
chr4_+_8633116
|
0.848
|
NM_080819
|
GPR78
|
G protein-coupled receptor 78
|
|
chr10_+_85944383
|
0.848
|
NM_001171971
NM_033100
|
CDHR1
|
cadherin-related family member 1
|
|
chr12_+_124115928
|
0.846
|
|
AACS
|
acetoacetyl-CoA synthetase
|
|
chr1_+_203106600
|
0.844
|
|
NFASC
|
neurofascin
|
|
chr12_-_131256525
|
0.842
|
NM_021808
|
GALNT9
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 9 (GalNAc-T9)
|
|
chr19_-_18910790
|
0.835
|
NM_001145724
|
HOMER3
|
homer homolog 3 (Drosophila)
|
|
chr21_-_32573195
|
0.833
|
NM_018944
|
MIS18A
|
MIS18 kinetochore protein homolog A (S. pombe)
|
|
chr5_-_179951071
|
0.830
|
NM_052863
|
SCGB3A1
|
secretoglobin, family 3A, member 1
|
|
chr2_+_220087135
|
0.826
|
NM_018674
NM_182847
|
ACCN4
|
amiloride-sensitive cation channel 4, pituitary
|
|
chr4_+_134289893
|
0.825
|
NM_020815
NM_032961
|
PCDH10
|
protocadherin 10
|
|
chr6_+_32240357
|
0.825
|
NM_030652
|
EGFL8
|
EGF-like-domain, multiple 8
|
|
chr19_-_59676153
|
0.825
|
NM_145057
|
CDC42EP5
|
CDC42 effector protein (Rho GTPase binding) 5
|
|
chr8_-_10550026
|
0.818
|
NM_178857
|
RP1L1
|
retinitis pigmentosa 1-like 1
|
|
chr17_-_1342700
|
0.818
|
NM_001080779
|
MYO1C
|
myosin IC
|