|
chr2_-_215967442
|
6.417
|
|
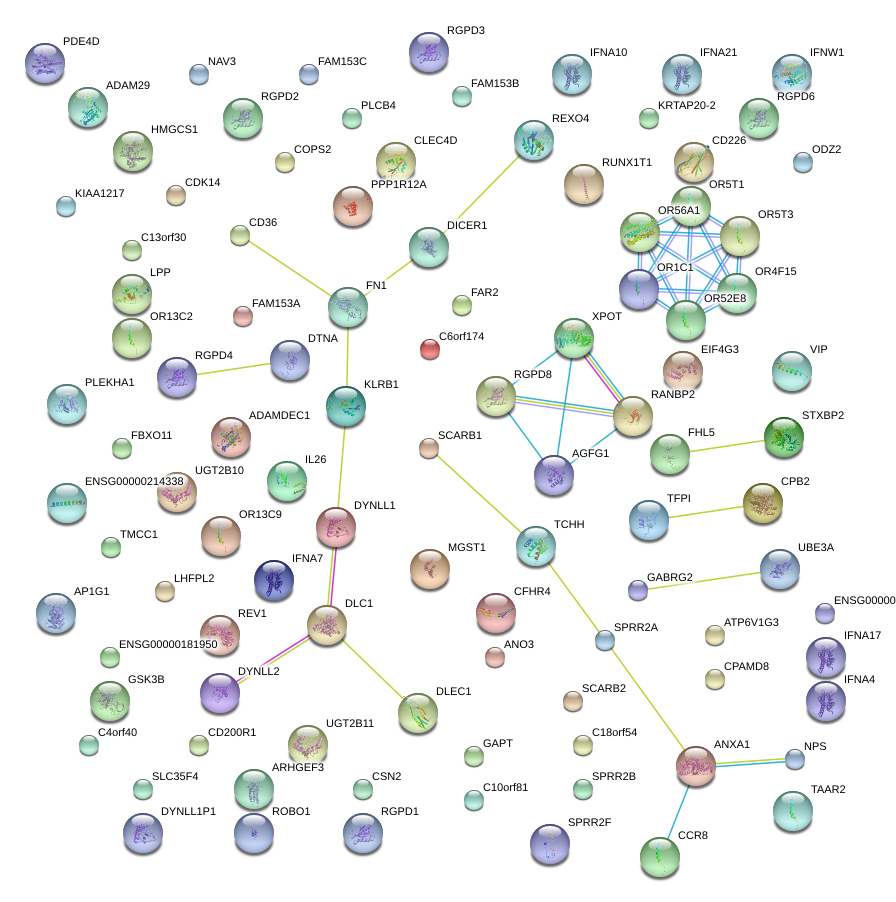
FN1
|
fibronectin 1
|
|
chr2_-_47969350
|
5.688
|
NM_025133
|
FBXO11
|
F-box protein 11
|
|
chr3_-_114176401
|
5.270
|
NM_138806
NM_138939
NM_138940
NM_170780
|
CD200R1
|
CD200 receptor 1
|
|
chr13_-_45577144
|
4.847
|
NM_001872
NM_016413
|
CPB2
|
carboxypeptidase B2 (plasma)
|
|
chr2_+_228064531
|
4.591
|
|
AGFG1
|
ArfGAP with FG repeats 1
|
|
chr21_-_26345294
|
4.449
|
|
|
|
|
chr9_-_21218132
|
4.381
|
NM_021268
|
IFNA17
|
interferon, alpha 17
|
|
chr3_-_56784634
|
4.301
|
NM_001128616
|
ARHGEF3
|
Rho guanine nucleotide exchange factor (GEF) 3
|
|
chr13_+_107753764
|
4.208
|
|
|
|
|
chr9_-_106407728
|
4.174
|
NM_001004481
|
OR13C2
|
olfactory receptor, family 13, subfamily C, member 2
|
|
chr9_-_21132143
|
4.044
|
NM_002177
|
IFNW1
|
interferon, omega 1
|
|
chr5_-_58607673
|
3.910
|
NM_001197220
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr12_+_16397617
|
3.798
|
NM_145764
|
MGST1
|
microsomal glutathione S-transferase 1
|
|
chr6_+_153113624
|
3.591
|
NM_003381
NM_194435
|
VIP
|
vasoactive intestinal peptide
|
|
chr4_+_176076083
|
3.510
|
NM_001130703
NM_001130704
NM_001130705
NM_014269
|
ADAM29
|
ADAM metallopeptidase domain 29
|
|
chr12_-_66905802
|
3.431
|
NM_018402
|
IL26
|
interleukin 26
|
|
chr2_+_108751033
|
3.402
|
|
RANBP2
|
RAN binding protein 2
|
|
chr1_-_21310462
|
3.381
|
NM_003760
|
EIF4G3
|
eukaryotic translation initiation factor 4 gamma, 3
|
|
chr18_-_65766068
|
3.275
|
|
CD226
|
CD226 molecule
|
|
chr11_+_55776251
|
3.218
|
NM_001004747
|
OR5T3
|
olfactory receptor, family 5, subfamily T, member 3
|
|
chr11_-_5835507
|
3.179
|
NM_001005168
|
OR52E8
|
olfactory receptor, family 52, subfamily E, member 8
|
|
chr11_+_55799604
|
3.150
|
NM_001004745
|
OR5T1
|
olfactory receptor, family 5, subfamily T, member 1
|
|
chr12_+_63090008
|
3.138
|
|
XPOT
|
exportin, tRNA (nuclear export receptor for tRNAs)
|
|
chr8_-_93099040
|
3.127
|
NM_175636
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr3_-_79899748
|
3.111
|
NM_002941
|
ROBO1
|
roundabout, axon guidance receptor, homolog 1 (Drosophila)
|
|
chr5_+_161427225
|
3.046
|
NM_000816
NM_198903
NM_198904
|
GABRG2
|
gamma-aminobutyric acid (GABA) A receptor, gamma 2
|
|
chr10_+_129237602
|
2.966
|
NM_001030013
|
NPS
|
neuropeptide S
|
|
chr14_-_57133361
|
2.933
|
NM_001080455
|
SLC35F4
|
solute carrier family 35, member F4
|
|
chr2_-_99412694
|
2.921
|
|
REV1
|
REV1 homolog (S. cerevisiae)
|
|
chr1_-_150353179
|
2.904
|
NM_007113
|
TCHH
|
trichohyalin
|
|
chr4_+_71054492
|
2.875
|
NM_214711
|
C4orf40
|
chromosome 4 open reading frame 40
|
|
chr13_+_42253685
|
2.856
|
NM_182508
|
C13orf30
|
chromosome 13 open reading frame 30
|
|
chr6_-_132987106
|
2.853
|
NM_001033080
|
TAAR2
|
trace amine associated receptor 2
|
|
chr1_-_245988330
|
2.844
|
NM_012353
|
OR1C1
|
olfactory receptor, family 1, subfamily C, member 1
|
|
chr7_+_90176647
|
2.839
|
NM_012395
|
CDK14
|
cyclin-dependent kinase 14
|
|
chr5_+_57823017
|
2.801
|
NM_152687
|
GAPT
|
GRB2-binding adaptor protein, transmembrane
|
|
chr4_-_70861301
|
2.779
|
NM_001891
|
CSN2
|
casein beta
|
|
chr2_-_95846109
|
2.754
|
|
LOC150759
|
hypothetical LOC150759
|
|
chr15_-_23204887
|
2.753
|
NM_000462
|
UBE3A
|
ubiquitin protein ligase E3A
|
|
chr12_+_29267863
|
2.731
|
NM_018099
|
FAR2
|
fatty acyl CoA reductase 2
|
|
chr4_+_69716298
|
2.676
|
NM_001075
NM_001144767
|
UGT2B10
|
UDP glucuronosyltransferase 2 family, polypeptide B10
|
|
chr9_+_74956601
|
2.673
|
|
ANXA1
|
annexin A1
|
|
chr9_-_21192201
|
2.650
|
NM_021057
|
IFNA7
|
interferon, alpha 7
|
|
chr14_-_94677807
|
2.642
|
NM_030621
|
DICER1
|
dicer 1, ribonuclease type III
|
|
chr8_-_13018123
|
2.635
|
NM_001164271
|
DLC1
|
deleted in liver cancer 1
|
|
chr2_-_188086612
|
2.628
|
|
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor)
|
|
chr7_+_80145461
|
2.618
|
|
CD36
|
CD36 molecule (thrombospondin receptor)
|
|
chr6_+_97117144
|
2.602
|
NM_001170807
NM_020482
|
FHL5
|
four and a half LIM domains 5
|
|
chr1_+_195123766
|
2.598
|
NM_006684
|
CFHR4
|
complement factor H-related 4
|
|
chr3_-_131081910
|
2.592
|
NM_001017395
|
TMCC1
|
transmembrane and coiled-coil domain family 1
|
|
chr8_+_24297742
|
2.586
|
NM_001145271
NM_001145272
NM_014479
|
ADAMDEC1
|
ADAM-like, decysin 1
|
|
chr21_+_30929425
|
2.573
|
NM_181616
|
KRTAP20-2
|
keratin associated protein 20-2
|
|
chr16_-_70339666
|
2.562
|
|
AP1G1
|
adaptor-related protein complex 1, gamma 1 subunit
|
|
chr11_-_22838547
|
2.550
|
NM_001195637
|
LOC100500938
|
hypothetical LOC100500938
|
|
chr12_-_78831821
|
2.525
|
NM_001143886
|
PPP1R12A
|
protein phosphatase 1, regulatory (inhibitor) subunit 12A
|
|
chr15_+_100175888
|
2.525
|
NM_001001674
|
OR4F15
|
olfactory receptor, family 4, subfamily F, member 15
|
|
chr3_+_189425886
|
2.501
|
NM_001167671
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr10_+_124141808
|
2.460
|
NM_021622
|
PLEKHA1
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1
|
|
chr3_-_121117647
|
2.454
|
|
GSK3B
|
glycogen synthase kinase 3 beta
|
|
chr18_+_30544193
|
2.439
|
NM_001198941
NM_032978
|
DTNA
|
dystrobrevin, alpha
|
|
chr12_+_8557402
|
2.375
|
NM_080387
|
CLEC4D
|
C-type lectin domain family 4, member D
|
|
chr5_-_43326922
|
2.368
|
|
HMGCS1
|
3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble)
|
|
chr12_+_76749199
|
2.354
|
NM_014903
|
NAV3
|
neuron navigator 3
|
|
chr1_-_151352587
|
2.327
|
NM_001014450
|
SPRR2A
SPRR2F
|
small proline-rich protein 2A
small proline-rich protein 2F
|
|
chr5_+_175444454
|
2.323
|
NM_001079529
|
FAM153B
|
family with sequence similarity 153, member B
|
|
chr10_+_24537725
|
2.321
|
NM_001098501
NM_019590
|
KIAA1217
|
KIAA1217
|
|
chr11_+_26310253
|
2.316
|
NM_031418
|
ANO3
|
anoctamin 3
|
|
chr5_+_166644420
|
2.303
|
NM_001122679
|
ODZ2
|
odz, odd Oz/ten-m homolog 2 (Drosophila)
|
|
chr6_-_127882192
|
2.287
|
NM_001012279
|
C6orf174
|
chromosome 6 open reading frame 174
|
|
chr3_+_39346200
|
2.286
|
NM_005201
|
CCR8
|
chemokine (C-C motif) receptor 8
|
|
chr18_+_50139168
|
2.280
|
NM_173529
|
C18orf54
|
chromosome 18 open reading frame 54
|
|
chr10_+_115514567
|
2.243
|
NM_182601
|
C10orf81
|
chromosome 10 open reading frame 81
|
|
chr5_-_77880731
|
2.229
|
|
LHFPL2
|
lipoma HMGIC fusion partner-like 2
|
|
chr20_+_9406460
|
2.228
|
|
PLCB4
|
phospholipase C, beta 4
|
|
chr11_-_6005546
|
2.227
|
NM_001001917
|
OR56A1
|
olfactory receptor, family 56, subfamily A, member 1
|
|
chr1_-_196776697
|
2.223
|
NM_133262
NM_133326
|
ATP6V1G3
|
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G3
|
|
chrX_+_118598325
|
2.210
|
NM_181777
|
UBE2A
|
ubiquitin-conjugating enzyme E2A (RAD6 homolog)
|
|
chr14_-_62638336
|
2.201
|
NM_172376
|
KCNH5
|
potassium voltage-gated channel, subfamily H (eag-related), member 5
|
|
chr11_-_55460451
|
2.185
|
NM_006637
|
OR5I1
|
olfactory receptor, family 5, subfamily I, member 1
|
|
chr4_+_157044296
|
2.183
|
NM_005651
|
TDO2
|
tryptophan 2,3-dioxygenase
|
|
chr3_-_193928038
|
2.178
|
NM_004113
|
FGF12
|
fibroblast growth factor 12
|
|
chr15_-_23201740
|
2.176
|
NM_130838
|
UBE3A
|
ubiquitin protein ligase E3A
|
|
chr9_-_21295311
|
2.175
|
NM_002169
|
IFNA5
|
interferon, alpha 5
|
|
chr11_-_55128424
|
2.156
|
NM_001004700
|
OR4C11
|
olfactory receptor, family 4, subfamily C, member 11
|
|
chr2_+_108230082
|
2.153
|
NM_001008743
|
SULT1C3
|
sulfotransferase family, cytosolic, 1C, member 3
|
|
chr8_+_118216517
|
2.151
|
NM_001172814
NM_173851
|
SLC30A8
|
solute carrier family 30 (zinc transporter), member 8
|
|
chr4_+_70896236
|
2.146
|
NM_001009181
NM_003154
|
STATH
|
statherin
|
|
chr9_-_103185621
|
2.145
|
NM_001127610
|
BAAT
|
bile acid CoA: amino acid N-acyltransferase (glycine N-choloyltransferase)
|
|
chr11_-_4827013
|
2.133
|
NM_001004758
|
OR51S1
|
olfactory receptor, family 51, subfamily S, member 1
|
|
chr18_-_29882555
|
2.124
|
NM_001198549
|
NOL4
|
nucleolar protein 4
|
|
chr18_+_59733724
|
2.120
|
NM_005024
|
SERPINB10
|
serpin peptidase inhibitor, clade B (ovalbumin), member 10
|
|
chr9_+_111927601
|
2.101
|
NM_001136562
|
AKAP2
|
A kinase (PRKA) anchor protein 2
|
|
chr6_+_127940011
|
2.098
|
NM_001010905
|
C6orf58
|
chromosome 6 open reading frame 58
|
|
chr9_+_6318348
|
2.094
|
NM_001001874
NM_001001875
NM_033516
|
TPD52L3
|
tumor protein D52-like 3
|
|
chr2_+_203901160
|
2.090
|
NM_005759
|
ABI2
|
abl-interactor 2
|
|
chr13_-_35686665
|
2.089
|
NM_017826
|
SOHLH2
|
spermatogenesis and oogenesis specific basic helix-loop-helix 2
|
|
chr9_-_14900992
|
2.089
|
NM_144966
|
FREM1
|
FRAS1 related extracellular matrix 1
|
|
chr5_+_179065282
|
2.035
|
|
CANX
|
calnexin
|
|
chr7_+_90177082
|
2.031
|
|
CDK14
|
cyclin-dependent kinase 14
|
|
chr4_+_91375204
|
2.028
|
NM_207491
|
FAM190A
|
family with sequence similarity 190, member A
|
|
chr11_-_62753574
|
2.027
|
NM_199352
|
SLC22A25
|
solute carrier family 22, member 25
|
|
chr19_+_41418723
|
2.024
|
|
ZNF146
|
zinc finger protein 146
|
|
chr13_+_108046500
|
2.019
|
NM_015011
|
MYO16
|
myosin XVI
|
|
chr21_-_30908093
|
2.012
|
NM_181602
|
KRTAP6-1
|
keratin associated protein 6-1
|
|
chr11_-_121492010
|
1.997
|
NM_001001786
|
BLID
|
BH3-like motif containing, cell death inducer
|
|
chr3_+_171112053
|
1.991
|
NM_182610
|
SAMD7
|
sterile alpha motif domain containing 7
|
|
chr8_+_107529327
|
1.985
|
NM_001198532
|
OXR1
|
oxidation resistance 1
|
|
chr12_+_46802695
|
1.983
|
NM_001166687
NM_001166688
|
PFKM
|
phosphofructokinase, muscle
|
|
chr5_-_41297296
|
1.965
|
NM_001115131
|
C6
|
complement component 6
|
|
chr1_+_146827470
|
1.958
|
NM_173638
|
NBPF15
|
neuroblastoma breakpoint family, member 15
|
|
chr12_+_9871343
|
1.957
|
NM_016523
|
KLRF1
|
killer cell lectin-like receptor subfamily F, member 1
|
|
chr13_+_51933228
|
1.947
|
|
CKAP2
|
cytoskeleton associated protein 2
|
|
chr2_-_210888010
|
1.940
|
NM_079420
|
MYL1
|
myosin, light chain 1, alkali; skeletal, fast
|
|
chr14_-_105681377
|
1.937
|
|
IGHV4-31
IGHG1
|
immunoglobulin heavy variable 4-31
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr12_+_54080480
|
1.928
|
NM_001005518
|
OR6C65
|
olfactory receptor, family 6, subfamily C, member 65
|
|
chr6_-_64087806
|
1.911
|
NM_001143940
NM_016571
|
LGSN
|
lengsin, lens protein with glutamine synthetase domain
|
|
chr11_-_5110383
|
1.910
|
NM_001005160
|
OR52A5
|
olfactory receptor, family 52, subfamily A, member 5
|
|
chr11_-_57728228
|
1.908
|
NM_001004459
|
OR1S2
|
olfactory receptor, family 1, subfamily S, member 2
|
|
chr4_+_71298187
|
1.908
|
NM_021225
|
PROL1
|
proline rich, lacrimal 1
|
|
chr9_-_95255694
|
1.896
|
NM_198841
|
FAM120AOS
|
family with sequence similarity 120A opposite strand
|
|
chr4_-_70115037
|
1.885
|
NM_001073
|
UGT2B11
|
UDP glucuronosyltransferase 2 family, polypeptide B11
|
|
chr9_+_74956614
|
1.855
|
|
ANXA1
|
annexin A1
|
|
chr8_-_12220195
|
1.853
|
NM_001195257
NM_001037804
|
LOC100133267
DEFB130
|
defensin, beta 130-like
defensin, beta 130
|
|
chr9_+_124477135
|
1.841
|
NM_001005234
|
OR1L3
|
olfactory receptor, family 1, subfamily L, member 3
|
|
chr10_+_118177371
|
1.839
|
NM_001011709
|
PNLIPRP3
|
pancreatic lipase-related protein 3
|
|
chr4_+_114190233
|
1.838
|
NM_001148
NM_020977
|
ANK2
|
ankyrin 2, neuronal
|
|
chr9_-_103187082
|
1.834
|
NM_001701
|
BAAT
|
bile acid CoA: amino acid N-acyltransferase (glycine N-choloyltransferase)
|
|
chr1_-_203320153
|
1.820
|
NM_203376
|
TMEM81
|
transmembrane protein 81
|
|
chr4_+_87070449
|
1.819
|
NM_031305
|
ARHGAP24
|
Rho GTPase activating protein 24
|
|
chr1_+_63022359
|
1.806
|
NM_032852
NM_178221
|
ATG4C
|
ATG4 autophagy related 4 homolog C (S. cerevisiae)
|
|
chr8_-_18710575
|
1.789
|
NM_206909
|
PSD3
|
pleckstrin and Sec7 domain containing 3
|
|
chr1_-_246823691
|
1.776
|
NM_001004693
|
OR2T10
|
olfactory receptor, family 2, subfamily T, member 10
|
|
chr8_-_114458557
|
1.775
|
NM_198124
|
CSMD3
|
CUB and Sushi multiple domains 3
|
|
chr1_-_151296611
|
1.774
|
NM_005988
|
SPRR2A
|
small proline-rich protein 2A
|
|
chr14_+_31100330
|
1.772
|
NM_025152
|
NUBPL
|
nucleotide binding protein-like
|
|
chr8_+_50987149
|
1.764
|
NM_018967
|
SNTG1
|
syntrophin, gamma 1
|
|
chr8_-_30121741
|
1.760
|
NM_001100916
|
MBOAT4
|
membrane bound O-acyltransferase domain containing 4
|
|
chr2_-_215951161
|
1.756
|
|
FN1
|
fibronectin 1
|
|
chr8_-_102005697
|
1.752
|
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide
|
|
chr2_-_158008763
|
1.751
|
NM_004288
|
CYTIP
|
cytohesin 1 interacting protein
|
|
chr3_+_115099007
|
1.746
|
NM_001172105
|
GRAMD1C
|
GRAM domain containing 1C
|
|
chr12_+_10015274
|
1.737
|
NM_138337
NM_201623
|
CLEC12A
|
C-type lectin domain family 12, member A
|
|
chr11_-_5130174
|
1.736
|
NM_012375
|
OR52A1
|
olfactory receptor, family 52, subfamily A, member 1
|
|
chr13_+_81162030
|
1.731
|
|
|
|
|
chr2_+_190710730
|
1.728
|
NM_001042521
|
C2orf88
|
chromosome 2 open reading frame 88
|
|
chr15_+_49988882
|
1.726
|
|
TMOD3
|
tropomodulin 3 (ubiquitous)
|
|
chr9_+_21399132
|
1.709
|
NM_002170
|
IFNA8
|
interferon, alpha 8
|
|
chr2_+_114390975
|
1.702
|
|
ACTR3
|
ARP3 actin-related protein 3 homolog (yeast)
|
|
chr6_-_53638464
|
1.698
|
NM_001003760
|
KLHL31
|
kelch-like 31 (Drosophila)
|
|
chr12_+_32546243
|
1.696
|
NM_139241
|
FGD4
|
FYVE, RhoGEF and PH domain containing 4
|
|
chr11_-_104399104
|
1.683
|
NM_001136109
NM_001136110
NM_001136112
NM_004347
|
CASP5
|
caspase 5, apoptosis-related cysteine peptidase
|
|
chr9_+_74956494
|
1.679
|
NM_000700
|
ANXA1
|
annexin A1
|
|
chr5_+_102493155
|
1.678
|
NM_015216
|
PPIP5K2
|
diphosphoinositol pentakisphosphate kinase 2
|
|
chr21_-_30614477
|
1.657
|
NM_203405
|
KRTAP26-1
|
keratin associated protein 26-1
|
|
chr1_+_185064654
|
1.653
|
NM_024420
|
PLA2G4A
|
phospholipase A2, group IVA (cytosolic, calcium-dependent)
|
|
chr9_-_89939719
|
1.651
|
NM_001166137
|
FAM75C2
|
family with sequence similarity 75, member C2
|
|
chr4_+_71126376
|
1.650
|
NM_152997
|
C4orf7
|
chromosome 4 open reading frame 7
|
|
chr14_+_67238355
|
1.643
|
NM_152443
|
RDH12
|
retinol dehydrogenase 12 (all-trans/9-cis/11-cis)
|
|
chr8_-_62764839
|
1.643
|
NM_001164750
NM_001164751
NM_001164752
NM_001164753
NM_020164
NM_032467
NM_032468
|
ASPH
|
aspartate beta-hydroxylase
|
|
chr4_+_53975597
|
1.638
|
|
FIP1L1
|
FIP1 like 1 (S. cerevisiae)
|
|
chr2_+_159533329
|
1.638
|
NM_001145909
NM_033394
|
TANC1
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1
|
|
chr6_+_64344307
|
1.612
|
|
PTP4A1
|
protein tyrosine phosphatase type IVA, member 1
|
|
chr2_-_55090759
|
1.608
|
NM_007008
|
RTN4
|
reticulon 4
|
|
chr5_-_160211623
|
1.603
|
NM_025153
|
ATP10B
|
ATPase, class V, type 10B
|
|
chr12_-_10846435
|
1.602
|
NM_023919
|
TAS2R7
|
taste receptor, type 2, member 7
|
|
chr10_-_127559841
|
1.600
|
NM_018180
|
DHX32
|
DEAH (Asp-Glu-Ala-His) box polypeptide 32
|
|
chr3_-_152173475
|
1.598
|
NM_001195794
NM_174878
|
CLRN1
|
clarin 1
|
|
chr12_+_18305740
|
1.594
|
NM_004570
|
PIK3C2G
|
phosphoinositide-3-kinase, class 2, gamma polypeptide
|
|
chr4_-_191185390
|
1.592
|
NM_001005217
|
FRG2B
FRG2
|
FSHD region gene 2 family, member B
FSHD region gene 2
|
|
chr1_+_149165438
|
1.582
|
NM_001145415
NM_012432
|
SETDB1
|
SET domain, bifurcated 1
|
|
chr20_-_33783409
|
1.578
|
|
RBM39
|
RNA binding motif protein 39
|
|
chr9_+_78305368
|
1.576
|
NM_001097636
|
GCNT1
|
glucosaminyl (N-acetyl) transferase 1, core 2
|
|
chrX_-_131401319
|
1.574
|
NM_018388
NM_133486
|
MBNL3
|
muscleblind-like 3 (Drosophila)
|
|
chr8_-_17797127
|
1.570
|
NM_147203
NM_201553
NM_004467
NM_201552
|
FGL1
|
fibrinogen-like 1
|
|
chr4_+_159350850
|
1.568
|
NM_018342
|
TMEM144
|
transmembrane protein 144
|
|
chr3_-_25654659
|
1.568
|
|
TOP2B
|
topoisomerase (DNA) II beta 180kDa
|
|
chr3_-_107070400
|
1.567
|
NM_170662
|
CBLB
|
Cas-Br-M (murine) ecotropic retroviral transforming sequence b
|
|
chr11_-_5819702
|
1.564
|
NM_001005167
|
OR52E6
|
olfactory receptor, family 52, subfamily E, member 6
|
|
chr3_-_191650291
|
1.555
|
NM_207316
|
TMEM207
|
transmembrane protein 207
|
|
chr10_+_5228756
|
1.549
|
NM_001818
|
AKR1C4
|
aldo-keto reductase family 1, member C4 (chlordecone reductase; 3-alpha hydroxysteroid dehydrogenase, type I; dihydrodiol dehydrogenase 4)
|
|
chr11_-_123182266
|
1.548
|
NM_001005325
|
OR6M1
|
olfactory receptor, family 6, subfamily M, member 1
|
|
chr7_+_28441758
|
1.537
|
NM_004904
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr4_-_104240413
|
1.536
|
NM_020139
|
BDH2
|
3-hydroxybutyrate dehydrogenase, type 2
|
|
chr8_+_36760999
|
1.529
|
NM_001031836
|
KCNU1
|
potassium channel, subfamily U, member 1
|
|
chr14_-_30927931
|
1.528
|
NM_015473
|
HEATR5A
|
HEAT repeat containing 5A
|
|
chr19_-_7009644
|
1.526
|
NM_001164425
|
MBD3L3
|
methyl-CpG-binding domain protein 3-like 3-like
|
|
chr4_+_71283383
|
1.522
|
NM_006685
|
SMR3B
|
submaxillary gland androgen regulated protein 3B
|
|
chr3_-_167037946
|
1.518
|
NM_000055
|
BCHE
|
butyrylcholinesterase
|
|
chrX_+_105298953
|
1.518
|
NM_001171020
NM_152423
|
MUM1L1
|
melanoma associated antigen (mutated) 1-like 1
|
|
chr4_+_146820805
|
1.514
|
NM_001080531
|
C4orf51
|
chromosome 4 open reading frame 51
|
|
chr7_+_104539548
|
1.509
|
|
MLL5
|
myeloid/lymphoid or mixed-lineage leukemia 5 (trithorax homolog, Drosophila)
|
|
chr20_+_12937616
|
1.508
|
NM_018327
|
SPTLC3
|
serine palmitoyltransferase, long chain base subunit 3
|
|
chr12_+_53927248
|
1.498
|
NM_001005490
|
OR6C74
|
olfactory receptor, family 6, subfamily C, member 74
|
|
chr18_-_49944986
|
1.496
|
|
MBD2
|
methyl-CpG binding domain protein 2
|
|
chr3_-_99236520
|
1.496
|
NM_001105580
|
GABRR3
|
gamma-aminobutyric acid (GABA) receptor, rho 3
|
|
chr22_-_39189369
|
1.495
|
|
MKL1
|
megakaryoblastic leukemia (translocation) 1
|
|
chr4_-_21155376
|
1.483
|
NM_001035004
|
KCNIP4
|
Kv channel interacting protein 4
|
|
chr13_-_32678142
|
1.481
|
NM_052851
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr3_+_98641143
|
1.478
|
NM_173655
|
EPHA6
|
EPH receptor A6
|
|
chr13_+_48178620
|
1.478
|
NM_020377
|
CYSLTR2
|
cysteinyl leukotriene receptor 2
|