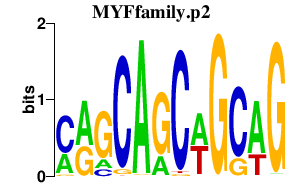
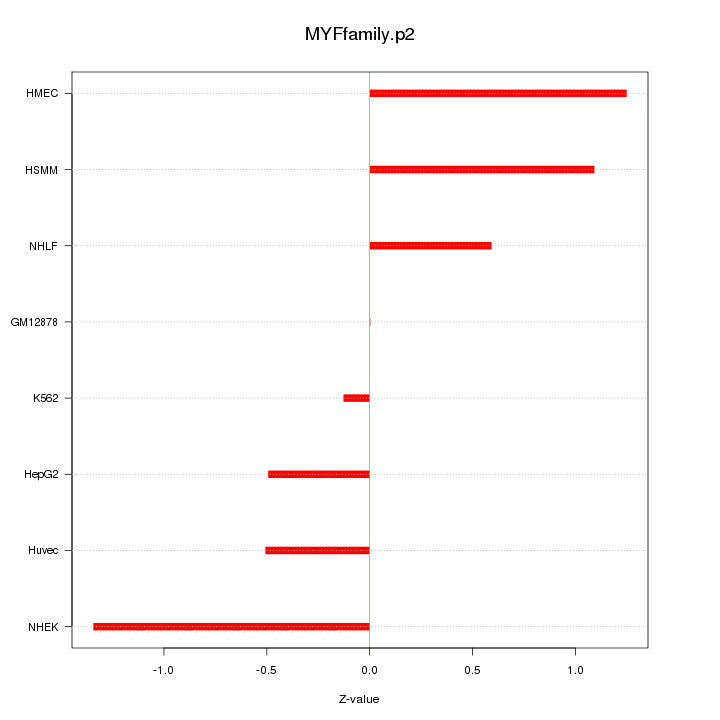
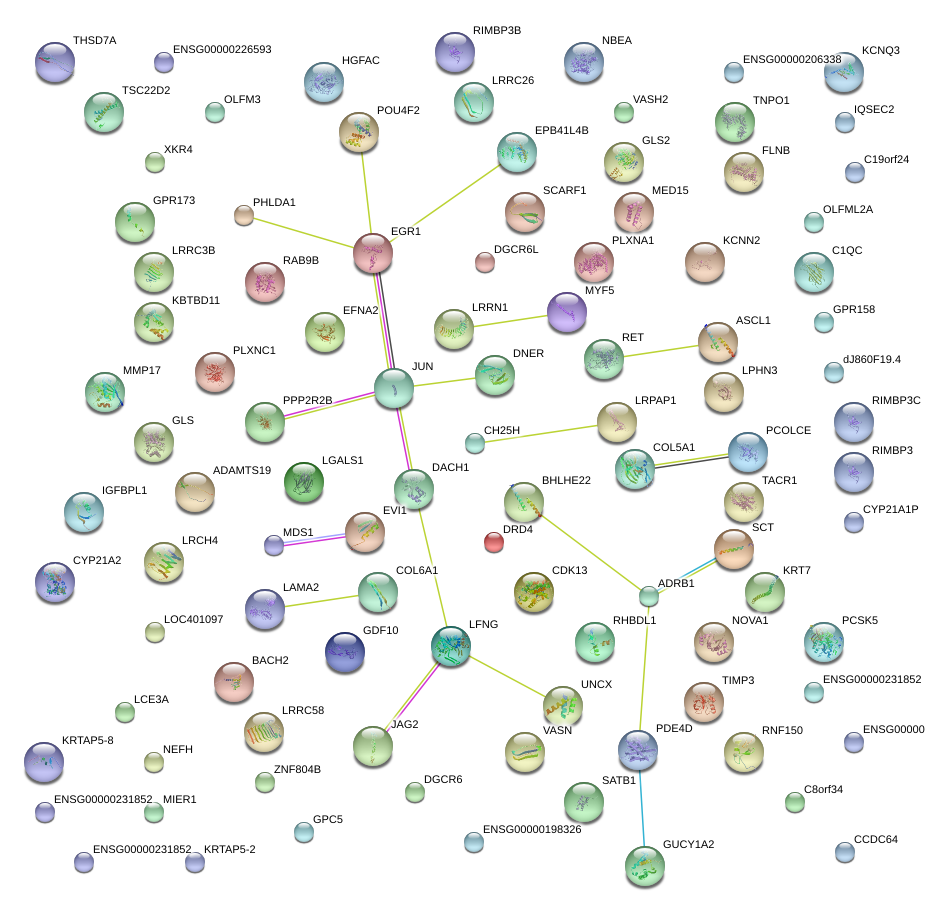
|
chr5_+_137829077
|
3.062
|
NM_001964
|
EGR1
|
early growth response 1
|
|
chr12_-_74710741
|
2.142
|
|
PHLDA1
|
pleckstrin homology-like domain, family A, member 1
|
|
chr5_+_113725914
|
1.867
|
NM_021614
|
KCNN2
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 2
|
|
chr5_+_137829065
|
1.857
|
|
EGR1
|
early growth response 1
|
|
chr8_+_56177567
|
1.569
|
NM_052898
|
XKR4
|
XK, Kell blood group complex subunit-related family, member 4
|
|
chr2_-_230287442
|
1.223
|
NM_139072
|
DNER
|
delta/notch-like EGF repeat containing
|
|
chr9_-_139183675
|
1.118
|
|
|
|
|
chr16_+_4370879
|
1.060
|
|
VASN
|
vasorin
|
|
chr3_+_3816079
|
1.007
|
NM_020873
|
LRRN1
|
leucine rich repeat neuronal 1
|
|
chr2_+_191453813
|
0.946
|
|
GLS
|
glutaminase
|
|
chr2_+_191453791
|
0.924
|
NM_014905
|
GLS
|
glutaminase
|
|
chr4_+_61749540
|
0.921
|
|
LPHN3
|
latrophilin 3
|
|
chr5_+_113726203
|
0.913
|
|
KCNN2
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 2
|
|
chr22_+_31527678
|
0.896
|
|
TIMP3
|
TIMP metallopeptidase inhibitor 3
|
|
chr9_-_139184247
|
0.885
|
NM_001013653
|
LRRC26
|
leucine rich repeat containing 26
|
|
chr3_+_128190070
|
0.879
|
NM_032242
|
PLXNA1
|
plexin A1
|
|
chr7_+_39956633
|
0.833
|
|
CDK13
|
cyclin-dependent kinase 13
|
|
chr5_-_59225377
|
0.832
|
NM_001104631
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr1_-_59020875
|
0.794
|
|
JUN
|
jun proto-oncogene
|
|
chr3_+_151608673
|
0.752
|
|
TSC22D2
|
TSC22 domain family, member 2
|
|
chr6_+_129245978
|
0.749
|
NM_000426
NM_001079823
|
LAMA2
|
laminin, alpha 2
|
|
chr1_-_150862202
|
0.733
|
NM_178431
|
LCE3A
|
late cornified envelope 3A
|
|
chr9_+_33667169
|
0.724
|
|
|
|
|
chr4_+_3413489
|
0.664
|
NM_001528
|
HGFAC
|
HGF activator
|
|
chr12_+_93066370
|
0.660
|
NM_005761
|
PLXNC1
|
plexin C1
|
|
chr7_-_11838260
|
0.649
|
NM_015204
|
THSD7A
|
thrombospondin, type I, domain containing 7A
|
|
chr3_+_161426322
|
0.647
|
|
LOC401097
|
hypothetical protein LOC401097
|
|
chr12_+_118911859
|
0.646
|
NM_207311
|
CCDC64
|
coiled-coil domain containing 64
|
|
chr3_-_18441763
|
0.634
|
NM_001195470
NM_002971
|
SATB1
|
SATB homeobox 1
|
|
chr2_+_191454036
|
0.632
|
|
GLS
|
glutaminase
|
|
chr14_-_26136799
|
0.625
|
NM_002515
NM_006489
NM_006491
|
NOVA1
|
neuro-oncological ventral antigen 1
|
|
chr10_+_42892506
|
0.620
|
NM_020630
NM_020975
|
RET
|
ret proto-oncogene
|
|
chrX_-_102973762
|
0.619
|
NM_016370
|
RAB9B
|
RAB9B, member RAS oncogene family
|
|
chr22_+_20068039
|
0.612
|
NM_001128635
|
RIMBP3B
|
RIMS binding protein 3B
|
|
chr22_+_28206180
|
0.609
|
NM_021076
|
NEFH
|
neurofilament, heavy polypeptide
|
|
chr2_+_191453792
|
0.602
|
|
GLS
|
glutaminase
|
|
chr13_+_34414390
|
0.601
|
NM_015678
|
NBEA
|
neurobeachin
|
|
chr21_+_46226072
|
0.590
|
NM_001848
|
COL6A1
|
collagen, type VI, alpha 1
|
|
chr13_-_71338144
|
0.587
|
|
DACH1
|
dachshund homolog 1 (Drosophila)
|
|
chr11_-_617172
|
0.573
|
NM_021920
|
SCT
|
secretin
|
|
chr22_+_31527744
|
0.558
|
|
TIMP3
|
TIMP metallopeptidase inhibitor 3
|
|
chr3_+_161425946
|
0.556
|
NM_001168214
|
LOC401097
|
hypothetical protein LOC401097
|
|
chr22_+_19248810
|
0.549
|
|
MED15
|
mediator complex subunit 15
|
|
chr4_+_3413457
|
0.546
|
|
HGFAC
|
HGF activator
|
|
chr12_+_50913220
|
0.538
|
NM_005556
|
KRT7
|
keratin 7
|
|
chr22_+_17273735
|
0.537
|
NM_005675
|
DGCR6
|
DiGeorge syndrome critical region gene 6
|
|
chr13_-_71338558
|
0.535
|
|
DACH1
|
dachshund homolog 1 (Drosophila)
|
|
chr19_+_1226510
|
0.529
|
NM_017914
|
C19orf24
|
chromosome 19 open reading frame 24
|
|
chr19_+_1236766
|
0.529
|
NM_001405
|
EFNA2
|
ephrin-A2
|
|
chr22_+_36401558
|
0.520
|
NM_002305
|
LGALS1
|
lectin, galactoside-binding, soluble, 1
|
|
chr7_+_39956480
|
0.517
|
NM_003718
NM_031267
|
CDK13
|
cyclin-dependent kinase 13
|
|
chr3_+_26639300
|
0.515
|
NM_052953
|
LRRC3B
|
leucine rich repeat containing 3B
|
|
chr12_+_50913274
|
0.506
|
|
KRT7
|
keratin 7
|
|
chr10_-_48058981
|
0.504
|
NM_004962
|
GDF10
|
growth differentiation factor 10
|
|
chr12_+_101876151
|
0.498
|
|
ASCL1
|
achaete-scute complex homolog 1 (Drosophila)
|
|
chr1_+_67163165
|
0.496
|
NM_001077700
NM_001077702
NM_001077703
NM_001146110
NM_001146111
NM_001146112
NM_001146113
NM_020948
|
MIER1
|
mesoderm induction early response 1 homolog (Xenopus laevis)
|
|
chr10_+_25504295
|
0.487
|
NM_020752
|
GPR158
|
G protein-coupled receptor 158
|
|
chr11_-_1576057
|
0.483
|
NM_001004325
|
KRTAP5-2
|
keratin associated protein 5-2
|
|
chr22_+_19248733
|
0.479
|
|
MED15
|
mediator complex subunit 15
|
|
chr14_-_104706159
|
0.479
|
NM_002226
NM_145159
|
JAG2
|
jagged 2
|
|
chr5_+_72179685
|
0.470
|
NM_153188
|
TNPO1
|
transportin 1
|
|
chrX_+_53095230
|
0.469
|
NM_018969
|
GPR173
|
G protein-coupled receptor 173
|
|
chr8_+_1937711
|
0.468
|
|
KBTBD11
|
kelch repeat and BTB (POZ) domain containing 11
|
|
chr13_+_90848875
|
0.465
|
NM_004466
|
GPC5
|
glypican 5
|
|
chr9_-_111122826
|
0.461
|
NM_018424
NM_019114
|
EPB41L4B
|
erythrocyte membrane protein band 4.1 like 4B
|
|
chr6_-_91063181
|
0.459
|
NM_021813
|
BACH2
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2
|
|
chr1_-_102234959
|
0.458
|
|
OLFM3
|
olfactomedin 3
|
|
chr11_+_627268
|
0.457
|
NM_000797
|
DRD4
|
dopamine receptor D4
|
|
chr2_-_75280121
|
0.456
|
NM_001058
NM_015727
|
TACR1
|
tachykinin receptor 1
|
|
chr5_-_146238336
|
0.456
|
NM_004576
NM_181675
NM_001127381
|
PPP2R2B
|
protein phosphatase 2, regulatory subunit B, beta
|
|
chr7_+_39956396
|
0.452
|
|
|
|
|
chr10_-_90957028
|
0.449
|
NM_003956
|
CH25H
|
cholesterol 25-hydroxylase
|
|
chr2_+_191454157
|
0.442
|
|
GLS
|
glutaminase
|
|
chr1_-_102235377
|
0.441
|
NM_058170
|
OLFM3
|
olfactomedin 3
|
|
chr9_-_111123017
|
0.441
|
|
EPB41L4B
|
erythrocyte membrane protein band 4.1 like 4B
|
|
chr8_-_133562185
|
0.441
|
NM_004519
|
KCNQ3
|
potassium voltage-gated channel, KQT-like subfamily, member 3
|
|
chr11_-_106394039
|
0.440
|
NM_000855
|
GUCY1A2
|
guanylate cyclase 1, soluble, alpha 2
|
|
chr3_-_121550767
|
0.433
|
NM_001099678
|
LRRC58
|
leucine rich repeat containing 58
|
|
chr9_+_126579203
|
0.429
|
NM_182487
|
OLFML2A
|
olfactomedin-like 2A
|
|
chr8_+_65656164
|
0.420
|
|
BHLHE22
|
basic helix-loop-helix family, member e22
|
|
chr1_+_22842704
|
0.420
|
NM_001114101
NM_172369
|
C1QC
|
complement component 1, q subcomponent, C chain
|
|
chr3_-_62835601
|
0.409
|
|
|
|
|
chrX_-_53367246
|
0.406
|
NM_001111125
|
IQSEC2
|
IQ motif and Sec7 domain 2
|
|
chr12_+_79634820
|
0.404
|
NM_005593
|
MYF5
|
myogenic factor 5
|
|
chr14_-_104705753
|
0.398
|
|
JAG2
|
jagged 2
|
|
chr7_+_2525922
|
0.396
|
NM_001040167
NM_001040168
|
LFNG
|
LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase
|
|
chr7_+_1239078
|
0.396
|
NM_001080461
|
UNCX
|
UNC homeobox
|
|
chr10_+_115793795
|
0.392
|
NM_000684
|
ADRB1
|
adrenergic, beta-1-, receptor
|
|
chr17_-_1495742
|
0.387
|
NM_003693
NM_145350
NM_145352
|
SCARF1
|
scavenger receptor class F, member 1
|
|
chr12_+_130878870
|
0.386
|
NM_016155
|
MMP17
|
matrix metallopeptidase 17 (membrane-inserted)
|
|
chr9_+_77695334
|
0.386
|
NM_001190482
NM_006200
|
PCSK5
|
proprotein convertase subtilisin/kexin type 5
|
|
chr8_+_69405510
|
0.382
|
NM_001195639
NM_052958
|
C8orf34
|
chromosome 8 open reading frame 34
|
|
chr1_+_211190631
|
0.379
|
|
VASH2
|
vasohibin 2
|
|
chr4_-_142273881
|
0.378
|
NM_020724
|
RNF150
|
ring finger protein 150
|
|
chr4_+_147779492
|
0.376
|
NM_004575
|
POU4F2
|
POU class 4 homeobox 2
|
|
chr6_+_32114060
|
0.373
|
NM_000500
NM_001128590
|
CYP21A2
|
cytochrome P450, family 21, subfamily A, polypeptide 2
|
|
chr7_+_100037817
|
0.371
|
NM_002593
|
PCOLCE
|
procollagen C-endopeptidase enhancer
|
|
chr9_+_136673534
|
0.371
|
|
COL5A1
|
collagen, type V, alpha 1
|
|
chr4_-_3503951
|
0.370
|
NM_002337
|
LRPAP1
|
low density lipoprotein receptor-related protein associated protein 1
|
|
chr9_-_38414422
|
0.370
|
NM_001007563
|
IGFBPL1
|
insulin-like growth factor binding protein-like 1
|
|
chr7_+_88226617
|
0.365
|
NM_181646
|
ZNF804B
|
zinc finger protein 804B
|
|
chr16_+_665680
|
0.360
|
|
RHBDL1
|
rhomboid, veinlet-like 1 (Drosophila)
|
|
chr5_+_128823904
|
0.360
|
NM_133638
|
ADAMTS19
|
ADAM metallopeptidase with thrombospondin type 1 motif, 19
|
|
chr3_-_170864111
|
0.356
|
NM_004991
|
MECOM
|
MDS1 and EVI1 complex locus
|
|
chr20_+_2743613
|
0.354
|
NM_080739
|
C20orf141
|
chromosome 20 open reading frame 141
|
|
chr3_+_142252890
|
0.351
|
|
SPSB4
|
splA/ryanodine receptor domain and SOCS box containing 4
|
|
chr20_-_61462960
|
0.350
|
NM_000744
|
CHRNA4
|
cholinergic receptor, nicotinic, alpha 4
|
|
chr3_+_85090721
|
0.349
|
NM_001167674
NM_001167675
|
CADM2
|
cell adhesion molecule 2
|
|
chr11_+_1607580
|
0.349
|
NM_001001480
|
KRTAP5-5
|
keratin associated protein 5-5
|
|
chr1_+_153317887
|
0.347
|
NM_004952
|
EFNA3
|
ephrin-A3
|
|
chr7_+_98084531
|
0.345
|
NM_002523
|
NPTX2
|
neuronal pentraxin II
|
|
chr2_+_26249463
|
0.343
|
NM_001168241
|
FAM59B
|
family with sequence similarity 59, member B
|
|
chr7_+_119700924
|
0.342
|
NM_012281
|
KCND2
|
potassium voltage-gated channel, Shal-related subfamily, member 2
|
|
chr20_+_42807901
|
0.339
|
NM_022358
|
KCNK15
|
potassium channel, subfamily K, member 15
|
|
chr1_-_24067347
|
0.338
|
NM_000147
|
FUCA1
|
fucosidase, alpha-L- 1, tissue
|
|
chr1_+_208472817
|
0.338
|
NM_019605
|
SERTAD4
|
SERTA domain containing 4
|
|
chr1_+_3531447
|
0.338
|
|
TPRG1L
|
tumor protein p63 regulated 1-like
|
|
chrX_-_20194831
|
0.337
|
|
RPS6KA3
|
ribosomal protein S6 kinase, 90kDa, polypeptide 3
|
|
chr19_+_46390916
|
0.336
|
NM_030622
|
CYP2S1
|
cytochrome P450, family 2, subfamily S, polypeptide 1
|
|
chr1_+_53697659
|
0.336
|
NM_033067
|
DMRTB1
|
DMRT-like family B with proline-rich C-terminal, 1
|
|
chrX_+_16874651
|
0.336
|
NM_001080975
NM_004726
|
REPS2
|
RALBP1 associated Eps domain containing 2
|
|
chr19_+_40451626
|
0.336
|
|
USF2
|
upstream transcription factor 2, c-fos interacting
|
|
chr5_+_72179749
|
0.335
|
|
TNPO1
|
transportin 1
|
|
chr2_+_114916368
|
0.335
|
NM_020868
|
DPP10
|
dipeptidyl-peptidase 10 (non-functional)
|
|
chr2_-_47650973
|
0.335
|
NM_022055
|
KCNK12
|
potassium channel, subfamily K, member 12
|
|
chr3_+_141136896
|
0.333
|
|
CLSTN2
|
calsyntenin 2
|
|
chrX_+_46318135
|
0.332
|
NM_019886
|
CHST7
|
carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 7
|
|
chr16_+_88696172
|
0.332
|
|
FAM157C
|
family with sequence similarity 157, member C
|
|
chr3_-_22389126
|
0.332
|
|
ZNF385D
|
zinc finger protein 385D
|
|
chr4_+_14613395
|
0.331
|
NM_001177381
NM_001177382
NM_001177383
NM_001177384
NM_182485
NM_182646
|
CPEB2
|
cytoplasmic polyadenylation element binding protein 2
|
|
chrX_+_119176494
|
0.330
|
NM_001099685
NM_032498
|
RHOXF2B
RHOXF2
|
Rhox homeobox family, member 2B
Rhox homeobox family, member 2
|
|
chr10_+_52504239
|
0.330
|
NM_006258
|
PRKG1
|
protein kinase, cGMP-dependent, type I
|
|
chr11_-_46099531
|
0.330
|
NM_001101802
NM_016621
|
PHF21A
|
PHD finger protein 21A
|
|
chr1_+_216586199
|
0.329
|
|
TGFB2
|
transforming growth factor, beta 2
|
|
chrX_+_21302317
|
0.328
|
NM_001168647
NM_001168648
NM_001168649
NM_014927
|
CNKSR2
|
connector enhancer of kinase suppressor of Ras 2
|
|
chr8_-_133561933
|
0.325
|
|
KCNQ3
|
potassium voltage-gated channel, KQT-like subfamily, member 3
|
|
chr1_+_44643513
|
0.323
|
NM_018150
|
RNF220
|
ring finger protein 220
|
|
chr4_-_90448157
|
0.321
|
NM_198281
|
GPRIN3
|
GPRIN family member 3
|
|
chr6_+_99389318
|
0.321
|
|
POU3F2
|
POU class 3 homeobox 2
|
|
chr19_-_54618410
|
0.321
|
NM_178449
|
PTH2
|
parathyroid hormone 2
|
|
chrX_-_99551926
|
0.319
|
NM_001105243
NM_001184880
NM_020766
|
PCDH19
|
protocadherin 19
|
|
chr1_-_67163001
|
0.319
|
NM_024763
NM_207014
|
WDR78
|
WD repeat domain 78
|
|
chr16_+_2807164
|
0.317
|
NM_006799
NM_144956
NM_144957
|
PRSS21
|
protease, serine, 21 (testisin)
|
|
chr13_-_109236897
|
0.317
|
NM_003749
|
IRS2
|
insulin receptor substrate 2
|
|
chr16_+_85101734
|
0.315
|
|
FOXF1
|
forkhead box F1
|
|
chr2_+_187266899
|
0.315
|
NM_177454
|
FAM171B
|
family with sequence similarity 171, member B
|
|
chr16_+_620932
|
0.313
|
NM_053284
|
WFIKKN1
|
WAP, follistatin/kazal, immunoglobulin, kunitz and netrin domain containing 1
|
|
chr16_+_85101605
|
0.312
|
NM_001451
|
FOXF1
|
forkhead box F1
|
|
chr14_+_99220349
|
0.312
|
NM_006668
|
CYP46A1
|
cytochrome P450, family 46, subfamily A, polypeptide 1
|
|
chr6_+_118335381
|
0.311
|
NM_001029858
|
SLC35F1
|
solute carrier family 35, member F1
|
|
chr7_-_104440707
|
0.309
|
|
LOC100216545
|
hypothetical LOC100216545
|
|
chr2_+_233029076
|
0.308
|
NM_001631
|
ALPI
|
alkaline phosphatase, intestinal
|
|
chr10_+_120957084
|
0.308
|
NM_005308
|
GRK5
|
G protein-coupled receptor kinase 5
|
|
chr6_+_107917965
|
0.305
|
NM_018013
|
SOBP
|
sine oculis binding protein homolog (Drosophila)
|
|
chr2_-_48836313
|
0.304
|
NM_000233
|
LHCGR
|
luteinizing hormone/choriogonadotropin receptor
|
|
chr2_+_54536780
|
0.300
|
NM_003128
|
SPTBN1
|
spectrin, beta, non-erythrocytic 1
|
|
chr1_+_24944493
|
0.299
|
|
CLIC4
|
chloride intracellular channel 4
|
|
chrX_-_84521052
|
0.299
|
|
POF1B
|
premature ovarian failure, 1B
|
|
chr5_-_148013908
|
0.298
|
NM_000870
|
HTR4
|
5-hydroxytryptamine (serotonin) receptor 4
|
|
chr17_-_7962531
|
0.297
|
NM_021628
|
ALOXE3
|
arachidonate lipoxygenase 3
|
|
chr16_+_2807228
|
0.297
|
|
PRSS21
|
protease, serine, 21 (testisin)
|
|
chr11_+_22171150
|
0.292
|
NM_001142649
NM_213599
|
ANO5
|
anoctamin 5
|
|
chr4_+_14614462
|
0.290
|
|
CPEB2
|
cytoplasmic polyadenylation element binding protein 2
|
|
chr15_+_65904994
|
0.286
|
NM_001031807
|
SKOR1
|
SKI family transcriptional corepressor 1
|
|
chr1_+_109810743
|
0.286
|
|
SYPL2
|
synaptophysin-like 2
|
|
chr11_+_65414677
|
0.286
|
|
CCDC85B
|
coiled-coil domain containing 85B
|
|
chr5_-_39400319
|
0.285
|
NM_001737
|
C9
|
complement component 9
|
|
chr1_+_157408000
|
0.285
|
NM_021189
NM_001127173
|
CADM3
|
cell adhesion molecule 3
|
|
chr1_+_78126787
|
0.285
|
NM_001172309
NM_144573
|
NEXN
|
nexilin (F actin binding protein)
|
|
chr4_-_90447966
|
0.285
|
|
GPRIN3
|
GPRIN family member 3
|
|
chr11_-_14870326
|
0.282
|
NM_024514
|
CYP2R1
|
cytochrome P450, family 2, subfamily R, polypeptide 1
|
|
chr3_-_64186151
|
0.280
|
NM_198859
|
PRICKLE2
|
prickle homolog 2 (Drosophila)
|
|
chrX_+_23262905
|
0.278
|
NM_173495
|
PTCHD1
|
patched domain containing 1
|
|
chr11_-_2136129
|
0.278
|
|
|
|
|
chr2_+_79593567
|
0.278
|
NM_001164883
NM_004389
|
CTNNA2
|
catenin (cadherin-associated protein), alpha 2
|
|
chr8_-_65873658
|
0.277
|
NM_004820
|
CYP7B1
|
cytochrome P450, family 7, subfamily B, polypeptide 1
|
|
chr1_+_3531410
|
0.276
|
NM_182752
|
TPRG1L
|
tumor protein p63 regulated 1-like
|
|
chr7_+_93861808
|
0.275
|
NM_000089
|
COL1A2
|
collagen, type I, alpha 2
|
|
chr1_-_150044365
|
0.275
|
NM_001004432
|
LINGO4
|
leucine rich repeat and Ig domain containing 4
|
|
chrX_-_20195062
|
0.274
|
|
RPS6KA3
|
ribosomal protein S6 kinase, 90kDa, polypeptide 3
|
|
chr21_+_33364420
|
0.273
|
|
OLIG1
|
oligodendrocyte transcription factor 1
|
|
chr1_-_1840564
|
0.265
|
NM_178545
|
TMEM52
|
transmembrane protein 52
|
|
chr8_-_53640520
|
0.265
|
NM_207413
|
FAM150A
|
family with sequence similarity 150, member A
|
|
chr13_+_38159172
|
0.264
|
NM_207361
|
FREM2
|
FRAS1 related extracellular matrix protein 2
|
|
chr7_+_99793561
|
0.264
|
NM_178238
|
PILRB
|
paired immunoglobin-like type 2 receptor beta
|
|
chr1_+_237616457
|
0.263
|
|
CHRM3
|
cholinergic receptor, muscarinic 3
|
|
chr7_-_137182125
|
0.262
|
NM_004717
|
DGKI
|
diacylglycerol kinase, iota
|
|
chr16_+_2461500
|
0.262
|
NM_006181
|
NTN3
|
netrin 3
|
|
chr12_-_110521407
|
0.261
|
|
ATXN2
|
ataxin 2
|
|
chr8_+_106400322
|
0.260
|
NM_012082
|
ZFPM2
|
zinc finger protein, multitype 2
|
|
chr7_-_122313646
|
0.260
|
|
CADPS2
|
Ca++-dependent secretion activator 2
|
|
chr8_-_140784416
|
0.259
|
NM_016601
|
KCNK9
|
potassium channel, subfamily K, member 9
|
|
chr9_-_103540038
|
0.259
|
|
GRIN3A
|
glutamate receptor, ionotropic, N-methyl-D-aspartate 3A
|
|
chrX_+_51503219
|
0.259
|
NM_018094
|
GSPT2
|
G1 to S phase transition 2
|
|
chrX_-_20194629
|
0.258
|
NM_004586
|
RPS6KA3
|
ribosomal protein S6 kinase, 90kDa, polypeptide 3
|
|
chr12_+_73219826
|
0.258
|
|
|
|
|
chr19_+_46390980
|
0.257
|
|
CYP2S1
|
cytochrome P450, family 2, subfamily S, polypeptide 1
|
|
chr19_+_5641270
|
0.256
|
NM_033643
|
RPL36
|
ribosomal protein L36
|
|
chr19_-_47190160
|
0.256
|
NM_152296
|
ATP1A3
|
ATPase, Na+/K+ transporting, alpha 3 polypeptide
|
|
chr8_+_143542378
|
0.254
|
NM_001702
|
BAI1
|
brain-specific angiogenesis inhibitor 1
|