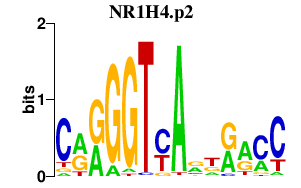
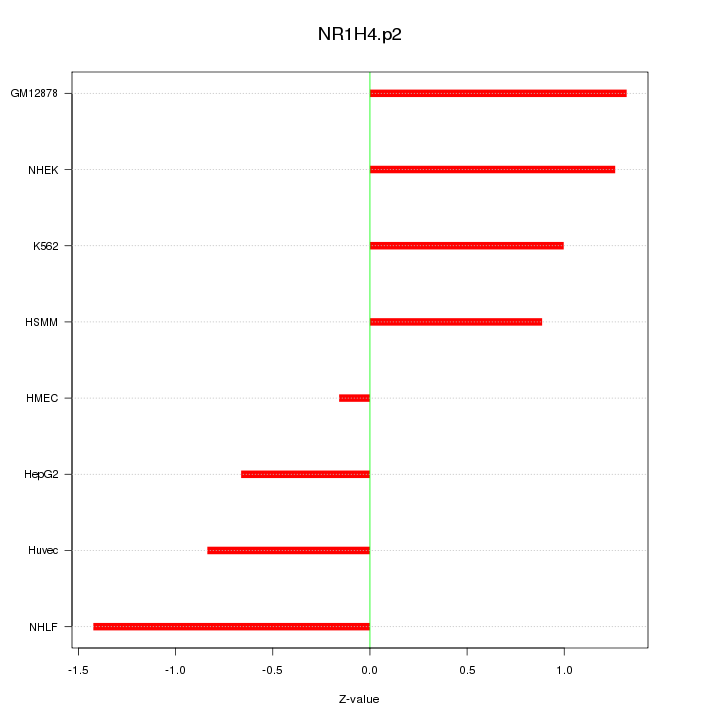
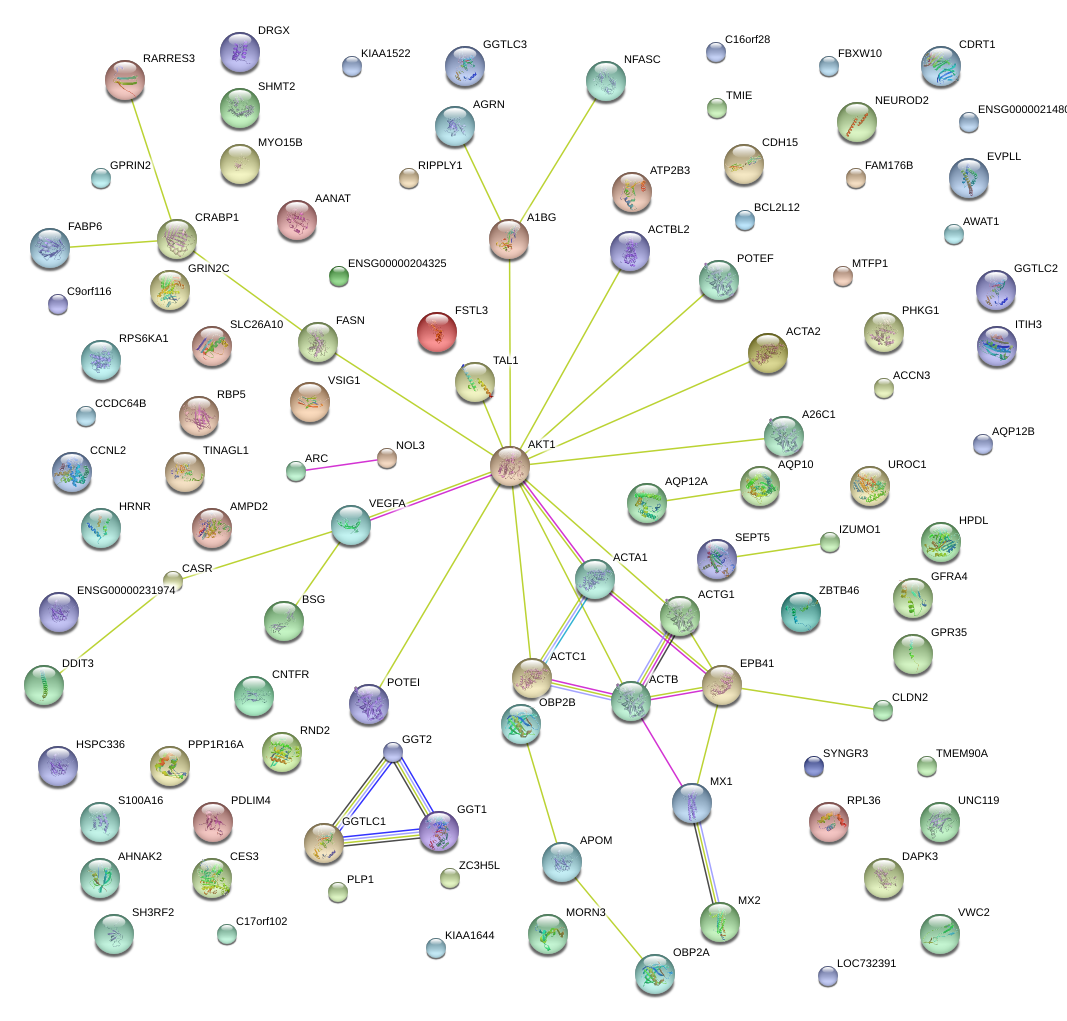
|
chr19_+_531666
|
5.207
|
|
BSG
|
basigin (Ok blood group)
|
|
chr1_+_93830169
|
3.733
|
|
LOC100129046
|
hypothetical LOC100129046
|
|
chr1_+_93830106
|
3.484
|
|
LOC100129046
|
hypothetical LOC100129046
|
|
chr1_-_47468029
|
2.349
|
NM_003189
|
TAL1
|
T-cell acute lymphocytic leukemia 1
|
|
chr1_+_33003847
|
2.113
|
|
KIAA1522
|
KIAA1522
|
|
chr1_+_31814672
|
2.062
|
NM_022164
|
TINAGL1
|
tubulointerstitial nephritis antigen-like 1
|
|
chr8_+_145692916
|
2.043
|
NM_032902
|
PPP1R16A
|
protein phosphatase 1, regulatory (inhibitor) subunit 16A
|
|
chr17_-_29930431
|
2.039
|
NM_207454
|
C17orf102
|
chromosome 17 open reading frame 102
|
|
chr1_+_31814712
|
1.987
|
|
TINAGL1
|
tubulointerstitial nephritis antigen-like 1
|
|
chr1_+_31814709
|
1.983
|
|
TINAGL1
|
tubulointerstitial nephritis antigen-like 1
|
|
chr20_-_61933012
|
1.974
|
|
ZBTB46
|
zinc finger and BTB domain containing 46
|
|
chr3_-_127719283
|
1.946
|
NM_001165974
NM_144639
|
UROC1
|
urocanase domain containing 1
|
|
chr3_+_52803823
|
1.759
|
NM_002217
|
ITIH3
|
inter-alpha (globulin) inhibitor H3
|
|
chr17_+_71975224
|
1.736
|
NM_001088
|
AANAT
|
aralkylamine N-acetyltransferase
|
|
chr1_+_33003814
|
1.723
|
NM_001198972
NM_001198973
|
KIAA1522
|
KIAA1522
|
|
chr16_+_87765661
|
1.701
|
NM_004933
|
CDH15
|
cadherin 15, type 1, M-cadherin (myotubule)
|
|
chr10_+_46413551
|
1.660
|
NM_014696
|
GPRIN2
|
G protein regulated inducer of neurite outgrowth 2
|
|
chr19_-_3920736
|
1.552
|
NM_001348
|
DAPK3
|
death-associated protein kinase 3
|
|
chr12_+_55909622
|
1.532
|
NM_001166356
NM_005412
|
SHMT2
|
serine hydroxymethyltransferase 2 (mitochondrial)
|
|
chr17_-_70367479
|
1.482
|
NM_000835
|
GRIN2C
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2C
|
|
chr19_-_53941835
|
1.465
|
NM_182575
|
IZUMO1
|
izumo sperm-egg fusion 1
|
|
chrX_+_102918453
|
1.441
|
|
PLP1
|
proteolipid protein 1
|
|
chr6_+_43845875
|
1.408
|
|
VEGFA
|
vascular endothelial growth factor A
|
|
chrX_-_106033202
|
1.398
|
NM_001171706
NM_138382
|
RIPPLY1
|
ripply1 homolog (zebrafish)
|
|
chr1_+_29113589
|
1.384
|
NM_001166006
|
EPB41
|
erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked)
|
|
chr19_+_627346
|
1.348
|
NM_005860
|
FSTL3
|
follistatin-like 3 (secreted glycoprotein)
|
|
chr17_-_40674883
|
1.333
|
|
|
|
|
chr17_+_18221803
|
1.322
|
NM_001145127
|
EVPLL
|
envoplakin-like
|
|
chr7_+_150376537
|
1.302
|
NM_004769
NM_020321
NM_020322
|
ACCN3
|
amiloride-sensitive cation channel 3
|
|
chr19_+_59657440
|
1.276
|
|
|
|
|
chr6_+_43846735
|
1.248
|
|
VEGFA
|
vascular endothelial growth factor A
|
|
chr14_-_73962557
|
1.241
|
NM_001105579
|
TMEM90A
|
transmembrane protein 90A
|
|
chr17_-_77647505
|
1.236
|
|
FASN
|
fatty acid synthase
|
|
chr9_-_137531517
|
1.231
|
NM_001048265
NM_144654
|
C9orf116
|
chromosome 9 open reading frame 116
|
|
chr1_+_109969571
|
1.224
|
NM_203404
|
AMPD2
|
adenosine monophosphate deaminase 2
|
|
chr1_+_152560179
|
1.207
|
NM_080429
|
AQP10
|
aquaporin 10
|
|
chr9_+_137577805
|
1.205
|
NM_014582
|
OBP2A
|
odorant binding protein 2A
|
|
chr1_+_559618
|
1.196
|
|
|
|
|
chr14_-_104507862
|
1.185
|
|
AHNAK2
|
AHNAK nucleoprotein 2
|
|
chr6_+_43846155
|
1.180
|
|
VEGFA
|
vascular endothelial growth factor A
|
|
chrX_+_152454773
|
1.137
|
NM_001001344
NM_021949
|
ATP2B3
|
ATPase, Ca++ transporting, plasma membrane 3
|
|
chr16_-_3025542
|
1.115
|
NM_001103175
|
CCDC64B
|
coiled-coil domain containing 64B
|
|
chr11_-_64133908
|
1.113
|
|
|
|
|
chr6_+_31731647
|
1.113
|
NM_019101
|
APOM
|
apolipoprotein M
|
|
chr6_+_31731667
|
1.107
|
|
APOM
|
apolipoprotein M
|
|
chr8_-_143692814
|
1.102
|
NM_015193
|
ARC
|
activity-regulated cytoskeleton-associated protein
|
|
chr3_+_46717826
|
1.096
|
NM_147196
|
TMIE
|
transmembrane inner ear
|
|
chr12_-_7172641
|
1.095
|
NM_031491
|
RBP5
|
retinol binding protein 5, cellular
|
|
chr1_+_26744893
|
1.081
|
|
RPS6KA1
|
ribosomal protein S6 kinase, 90kDa, polypeptide 1
|
|
chr5_+_145297490
|
1.070
|
|
SH3RF2
|
SH3 domain containing ring finger 2
|
|
chr17_+_38430773
|
1.070
|
NM_005440
|
RND2
|
Rho family GTPase 2
|
|
chr6_-_169392771
|
1.057
|
|
|
|
|
chr5_+_159589014
|
1.057
|
NM_001445
|
FABP6
|
fatty acid binding protein 6, ileal
|
|
chr9_-_34554800
|
1.056
|
|
CNTFR
|
ciliary neurotrophic factor receptor
|
|
chr16_+_1979959
|
1.051
|
NM_004209
|
SYNGR3
|
synaptogyrin 3
|
|
chr2_+_241213334
|
1.047
|
NM_005301
|
GPR35
|
G protein-coupled receptor 35
|
|
chr6_+_43846372
|
1.044
|
|
VEGFA
|
vascular endothelial growth factor A
|
|
chr5_+_145296313
|
1.043
|
NM_152550
|
SH3RF2
|
SH3 domain containing ring finger 2
|
|
chr6_+_43845915
|
1.040
|
NM_001025366
NM_001025367
NM_001025368
NM_001025369
NM_001025370
NM_001033756
NM_001171622
NM_001171623
NM_001171624
NM_001171625
NM_001171626
NM_001171627
NM_001171628
NM_001171629
NM_001171630
NM_003376
|
VEGFA
|
vascular endothelial growth factor A
|
|
chrX_+_69371229
|
1.038
|
NM_001013579
|
AWAT1
|
acyl-CoA wax alcohol acyltransferase 1
|
|
chr10_-_50269912
|
1.035
|
NM_001080520
|
DRGX
|
dorsal root ganglia homeobox
|
|
chr1_+_203179976
|
1.035
|
NM_001160331
|
NFASC
|
neurofascin
|
|
chr17_+_71097147
|
1.023
|
|
MYO15B
|
myosin XVB pseudogene
|
|
chr19_+_5641307
|
1.020
|
|
RPL36
|
ribosomal protein L36
|
|
chr11_-_64266916
|
1.017
|
|
|
|
|
chr14_-_104330982
|
1.014
|
NM_005163
|
AKT1
|
v-akt murine thymoma viral oncogene homolog 1
|
|
chr17_-_35017650
|
1.009
|
|
NEUROD2
|
neurogenic differentiation 2
|
|
chr21_+_41663853
|
0.993
|
|
MX2
|
myxovirus (influenza virus) resistance 2 (mouse)
|
|
chrX_+_102918409
|
0.984
|
NM_000533
NM_199478
|
PLP1
|
proteolipid protein 1
|
|
chr17_-_77092831
|
0.971
|
|
ACTG1
|
actin, gamma 1
|
|
chr19_-_63556632
|
0.971
|
NM_130786
|
A1BG
|
alpha-1-B glycoprotein
|
|
chr2_-_241270982
|
0.967
|
NM_001102467
|
AQP12A
AQP12B
|
aquaporin 12A
aquaporin 12B
|
|
chr12_-_56200547
|
0.965
|
NM_001195053
NM_001195054
NM_001195055
NM_001195056
NM_001195057
NM_004083
|
DDIT3
|
DNA-damage-inducible transcript 3
|
|
chr1_+_968522
|
0.961
|
|
AGRN
|
agrin
|
|
chr7_+_49783774
|
0.958
|
NM_198570
|
VWC2
|
von Willebrand factor C domain containing 2
|
|
chr12_+_56299959
|
0.942
|
NM_133489
|
SLC26A10
|
solute carrier family 26, member 10
|
|
chr16_-_1369613
|
0.936
|
NM_001193389
NM_023076
|
UNKL
|
unkempt homolog (Drosophila)-like
|
|
chr5_+_131621287
|
0.934
|
|
PDLIM4
|
PDZ and LIM domain 4
|
|
chr17_+_18588050
|
0.929
|
NM_031456
|
FBXW10
CDRT1
|
F-box and WD repeat domain containing 10
CMT1A duplicated region transcript 1
|
|
chr1_+_1324762
|
0.927
|
|
LOC148413
|
hypothetical LOC148413
|
|
chr16_+_65552600
|
0.923
|
NM_001185177
NM_024922
|
CES3
|
carboxylesterase 3
|
|
chr4_-_1185144
|
0.922
|
|
LOC100130872
|
hypothetical LOC100130872
|
|
chrX_+_107174855
|
0.920
|
NM_001170553
NM_182607
|
VSIG1
|
V-set and immunoglobulin domain containing 1
|
|
chr20_-_3592045
|
0.914
|
NM_022139
NM_145762
|
GFRA4
|
GDNF family receptor alpha 4
|
|
chrX_+_106030049
|
0.904
|
NM_001171092
|
CLDN2
|
claudin 2
|
|
chr1_-_36562341
|
0.899
|
NM_018166
|
FAM176B
|
family with sequence similarity 176, member B
|
|
chr1_+_45565126
|
0.896
|
NM_032756
|
HPDL
|
4-hydroxyphenylpyruvate dioxygenase-like
|
|
chr17_-_23903772
|
0.896
|
NM_005148
NM_054035
|
UNC119
|
unc-119 homolog (C. elegans)
|
|
chr1_-_151852128
|
0.894
|
NM_080388
|
S100A16
|
S100 calcium binding protein A16
|
|
chr1_-_1324511
|
0.892
|
NM_001039577
NM_030937
|
CCNL2
|
cyclin L2
|
|
chr19_+_5641332
|
0.885
|
NM_015414
|
RPL36
|
ribosomal protein L36
|
|
chr11_+_63060848
|
0.885
|
NM_004585
|
RARRES3
|
retinoic acid receptor responder (tazarotene induced) 3
|
|
chr22_-_43040063
|
0.883
|
NM_001099294
|
KIAA1644
|
KIAA1644
|
|
chr22_+_29151651
|
0.882
|
NM_001003704
NM_016498
|
MTFP1
|
mitochondrial fission process 1
|
|
chr19_+_5641270
|
0.879
|
NM_033643
|
RPL36
|
ribosomal protein L36
|
|
chr22_+_18085891
|
0.877
|
|
SEPT5
|
septin 5
|
|
chr22_+_23309717
|
0.869
|
NM_013430
|
GGT1
|
gamma-glutamyltransferase 1
|
|
chr22_+_28446288
|
0.865
|
NM_182527
|
CABP7
|
calcium binding protein 7
|
|
chr19_+_40895669
|
0.862
|
NM_014383
|
ZBTB32
|
zinc finger and BTB domain containing 32
|
|
chr12_+_50592379
|
0.858
|
NM_001077401
|
ACVRL1
|
activin A receptor type II-like 1
|
|
chr5_+_131621276
|
0.857
|
|
PDLIM4
|
PDZ and LIM domain 4
|
|
chr5_+_149549712
|
0.856
|
NM_014228
|
SLC6A7
|
solute carrier family 6 (neurotransmitter transporter, L-proline), member 7
|
|
chr14_-_105163301
|
0.855
|
|
IGHG4
|
immunoglobulin heavy constant gamma 4 (G4m marker)
|
|
chr1_+_26744929
|
0.851
|
NM_001006665
|
RPS6KA1
|
ribosomal protein S6 kinase, 90kDa, polypeptide 1
|
|
chrX_+_67965517
|
0.845
|
NM_004429
|
EFNB1
|
ephrin-B1
|
|
chr1_-_25128951
|
0.842
|
|
RUNX3
|
runt-related transcription factor 3
|
|
chr3_-_127810087
|
0.837
|
NM_001039783
|
TXNRD3IT1
|
thioredoxin reductase 3 intronic transcript 1
|
|
chr17_-_23903704
|
0.834
|
|
UNC119
|
unc-119 homolog (C. elegans)
|
|
chr5_+_133478252
|
0.830
|
NM_003202
|
TCF7
|
transcription factor 7 (T-cell specific, HMG-box)
|
|
chr1_-_25129354
|
0.823
|
NM_004350
|
RUNX3
|
runt-related transcription factor 3
|
|
chr19_+_17691283
|
0.822
|
|
MAP1S
|
microtubule-associated protein 1S
|
|
chr11_+_67007080
|
0.822
|
NM_003977
|
AIP
|
aryl hydrocarbon receptor interacting protein
|
|
chr22_+_18085990
|
0.819
|
|
SEPT5-GP1BB
SEPT5
|
SEPT5-GP1BB read-through transcript
septin 5
|
|
chr3_-_48607596
|
0.810
|
NM_000094
|
COL7A1
|
collagen, type VII, alpha 1
|
|
chr2_-_182253457
|
0.807
|
NM_002500
|
NEUROD1
|
neurogenic differentiation 1
|
|
chr17_-_35017691
|
0.804
|
NM_006160
|
NEUROD2
|
neurogenic differentiation 2
|
|
chr17_+_75776193
|
0.800
|
NM_052819
|
CARD14
|
caspase recruitment domain family, member 14
|
|
chr17_+_73676219
|
0.799
|
NM_004710
|
SYNGR2
|
synaptogyrin 2
|
|
chr19_-_50355247
|
0.797
|
NM_198478
|
NKPD1
|
NTPase, KAP family P-loop domain containing 1
|
|
chr2_-_218575922
|
0.796
|
|
TNS1
|
tensin 1
|
|
chr16_-_65775326
|
0.794
|
NM_001040715
|
KIAA0895L
|
KIAA0895-like
|
|
chr11_+_66381451
|
0.784
|
NM_024036
|
LRFN4
|
leucine rich repeat and fibronectin type III domain containing 4
|
|
chr16_+_66131247
|
0.781
|
|
FAM65A
|
family with sequence similarity 65, member A
|
|
chr4_-_69852090
|
0.780
|
NM_024743
|
UGT2A3
|
UDP glucuronosyltransferase 2 family, polypeptide A3
|
|
chr1_+_29086156
|
0.779
|
NM_001166005
NM_001166007
NM_004437
NM_203342
NM_203343
|
EPB41
|
erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked)
|
|
chr3_-_58627509
|
0.779
|
NM_138805
|
FAM3D
|
family with sequence similarity 3, member D
|
|
chrX_+_65152004
|
0.779
|
|
|
|
|
chr1_+_46541913
|
0.772
|
NM_006004
|
UQCRH
|
ubiquinol-cytochrome c reductase hinge protein
|
|
chr22_+_19651446
|
0.769
|
NM_001146288
|
AIFM3
|
apoptosis-inducing factor, mitochondrion-associated, 3
|
|
chr11_+_74539679
|
0.767
|
NM_001145212
NM_007256
|
SLCO2B1
|
solute carrier organic anion transporter family, member 2B1
|
|
chr17_+_45706786
|
0.765
|
NM_153229
|
TMEM92
|
transmembrane protein 92
|
|
chr16_-_2276875
|
0.763
|
|
|
|
|
chr9_+_35032222
|
0.763
|
NM_001040410
NM_203299
|
C9orf131
|
chromosome 9 open reading frame 131
|
|
chr16_+_30102231
|
0.763
|
NM_001193333
NM_007074
|
CORO1A
|
coronin, actin binding protein, 1A
|
|
chr9_+_126094388
|
0.761
|
NM_001166169
|
NEK6
|
NIMA (never in mitosis gene a)-related kinase 6
|
|
chr15_+_72695248
|
0.757
|
|
CLK3
|
CDC-like kinase 3
|
|
chr1_+_15637521
|
0.753
|
NM_007272
|
CTRC
|
chymotrypsin C (caldecrin)
|
|
chr1_+_29086206
|
0.752
|
|
EPB41
|
erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked)
|
|
chrX_+_102918089
|
0.750
|
NM_001128834
|
PLP1
|
proteolipid protein 1
|
|
chr7_+_2638156
|
0.748
|
|
TTYH3
|
tweety homolog 3 (Drosophila)
|
|
chr16_+_11669736
|
0.745
|
NM_003498
|
SNN
|
stannin
|
|
chr5_-_177355848
|
0.743
|
NM_006261
|
PROP1
|
PROP paired-like homeobox 1
|
|
chrX_-_21686079
|
0.739
|
NM_014332
|
SMPX
|
small muscle protein, X-linked
|
|
chr11_+_67007155
|
0.737
|
|
AIP
|
aryl hydrocarbon receptor interacting protein
|
|
chr19_+_17691302
|
0.735
|
NM_018174
|
MAP1S
|
microtubule-associated protein 1S
|
|
chr1_+_20338405
|
0.735
|
NM_022819
|
PLA2G2F
|
phospholipase A2, group IIF
|
|
chr1_+_17121012
|
0.722
|
NM_014675
|
CROCC
|
ciliary rootlet coiled-coil, rootletin
|
|
chr1_+_12046020
|
0.720
|
NM_001243
|
TNFRSF8
|
tumor necrosis factor receptor superfamily, member 8
|
|
chr19_-_47638890
|
0.717
|
NM_198477
|
CXCL17
|
chemokine (C-X-C motif) ligand 17
|
|
chr11_-_62136778
|
0.716
|
NM_153265
|
EML3
|
echinoderm microtubule associated protein like 3
|
|
chr11_+_118561375
|
0.714
|
NM_001168468
NM_024791
|
PDZD3
|
PDZ domain containing 3
|
|
chr5_+_131621230
|
0.712
|
NM_001131027
NM_003687
|
PDLIM4
|
PDZ and LIM domain 4
|
|
chr7_+_2638108
|
0.705
|
NM_025250
|
TTYH3
|
tweety homolog 3 (Drosophila)
|
|
chr17_-_76827370
|
0.702
|
|
C17orf56
|
chromosome 17 open reading frame 56
|
|
chr11_+_62136788
|
0.695
|
NM_000327
|
ROM1
|
retinal outer segment membrane protein 1
|
|
chr19_-_16497213
|
0.692
|
|
CHERP
|
calcium homeostasis endoplasmic reticulum protein
|
|
chr10_+_71060008
|
0.692
|
NM_145306
|
C10orf35
|
chromosome 10 open reading frame 35
|
|
chr6_+_41622267
|
0.690
|
|
FOXP4
|
forkhead box P4
|
|
chr11_+_64114857
|
0.687
|
NM_144585
NM_153378
|
SLC22A12
|
solute carrier family 22 (organic anion/urate transporter), member 12
|
|
chr15_+_70734091
|
0.685
|
NM_018652
|
GOLGA6A
GOLGA6B
|
golgin A6 family, member A
golgin A6 family, member B
|
|
chr22_+_17337970
|
0.683
|
|
DGCR5
|
DiGeorge syndrome critical region gene 5 (non-protein coding)
|
|
chr19_+_10424581
|
0.683
|
NM_006202
|
PDE4A
|
phosphodiesterase 4A, cAMP-specific
|
|
chr16_+_30102426
|
0.678
|
|
CORO1A
|
coronin, actin binding protein, 1A
|
|
chr22_-_37211467
|
0.672
|
|
DDX17
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 17
|
|
chr1_+_16131633
|
0.671
|
|
|
|
|
chr1_-_39871226
|
0.669
|
|
HEYL
|
hairy/enhancer-of-split related with YRPW motif-like
|
|
chr15_-_38185578
|
0.666
|
NM_001003942
NM_001003943
|
BMF
|
Bcl2 modifying factor
|
|
chr7_+_45899080
|
0.666
|
|
IGFBP1
|
insulin-like growth factor binding protein 1
|
|
chr1_+_32512298
|
0.665
|
NM_001042771
|
LCK
|
lymphocyte-specific protein tyrosine kinase
|
|
chr17_-_42250961
|
0.662
|
NM_030753
|
WNT3
|
wingless-type MMTV integration site family, member 3
|
|
chr13_+_113587674
|
0.660
|
|
GAS6
|
growth arrest-specific 6
|
|
chrX_+_151991474
|
0.659
|
NM_001170944
NM_032882
|
LOC100287428
PNMA6A
|
paraneoplastic antigen-like protein 6A related gene
paraneoplastic antigen like 6A
|
|
chr16_+_29892714
|
0.652
|
NM_004783
NM_016151
|
TAOK2
|
TAO kinase 2
|
|
chr19_+_40299058
|
0.652
|
|
FXYD3
|
FXYD domain containing ion transport regulator 3
|
|
chr1_-_154205675
|
0.648
|
|
ARHGEF2
|
Rho/Rac guanine nucleotide exchange factor (GEF) 2
|
|
chr17_+_71229191
|
0.646
|
|
ITGB4
|
integrin, beta 4
|
|
chr1_-_77457664
|
0.644
|
NM_005482
|
PIGK
|
phosphatidylinositol glycan anchor biosynthesis, class K
|
|
chr16_+_670426
|
0.644
|
|
STUB1
|
STIP1 homology and U-box containing protein 1, E3 ubiquitin protein ligase
|
|
chr11_+_65140207
|
0.644
|
NM_032223
|
PCNXL3
|
pecanex-like 3 (Drosophila)
|
|
chr17_-_7047285
|
0.641
|
|
DLG4
|
discs, large homolog 4 (Drosophila)
|
|
chr1_+_32485404
|
0.638
|
NM_032648
|
FAM167B
|
family with sequence similarity 167, member B
|
|
chr16_+_273118
|
0.638
|
NM_006849
|
PDIA2
|
protein disulfide isomerase family A, member 2
|
|
chr17_-_43390108
|
0.637
|
NM_024320
|
PRR15L
|
proline rich 15-like
|
|
chrX_+_151633730
|
0.636
|
NM_005361
NM_175742
NM_175743
|
MAGEA2
|
melanoma antigen family A, 2
|
|
chr17_+_76827699
|
0.630
|
NM_001086521
|
C17orf89
|
chromosome 17 open reading frame 89
|
|
chr12_-_51514328
|
0.630
|
NM_175834
|
KRT79
|
keratin 79
|
|
chr1_-_25129305
|
0.628
|
|
RUNX3
|
runt-related transcription factor 3
|
|
chr20_+_30192968
|
0.626
|
|
TM9SF4
|
transmembrane 9 superfamily protein member 4
|
|
chr6_-_33016561
|
0.624
|
NM_002118
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta
|
|
chr18_-_65775139
|
0.623
|
NM_006566
|
CD226
|
CD226 molecule
|
|
chr17_-_15463742
|
0.621
|
NM_006382
|
CDRT1
|
CMT1A duplicated region transcript 1
|
|
chr1_+_160618143
|
0.618
|
NM_001085375
|
C1orf226
|
chromosome 1 open reading frame 226
|
|
chr3_-_46693902
|
0.617
|
NM_182775
|
ALS2CL
|
ALS2 C-terminal like
|
|
chr1_-_16812274
|
0.612
|
NM_017940
|
NBPF1
|
neuroblastoma breakpoint family, member 1
|
|
chr17_+_77261447
|
0.611
|
|
HGS
|
hepatocyte growth factor-regulated tyrosine kinase substrate
|
|
chr11_-_3649120
|
0.608
|
NM_020402
|
CHRNA10
|
cholinergic receptor, nicotinic, alpha 10
|
|
chr6_+_44234525
|
0.605
|
NM_007058
|
CAPN11
|
calpain 11
|
|
chr2_+_27225362
|
0.603
|
NM_175769
|
TCF23
|
transcription factor 23
|
|
chr6_+_42231121
|
0.603
|
NM_000409
|
GUCA1A
|
guanylate cyclase activator 1A (retina)
|
|
chr9_+_137532294
|
0.603
|
NM_016034
|
MRPS2
|
mitochondrial ribosomal protein S2
|