|
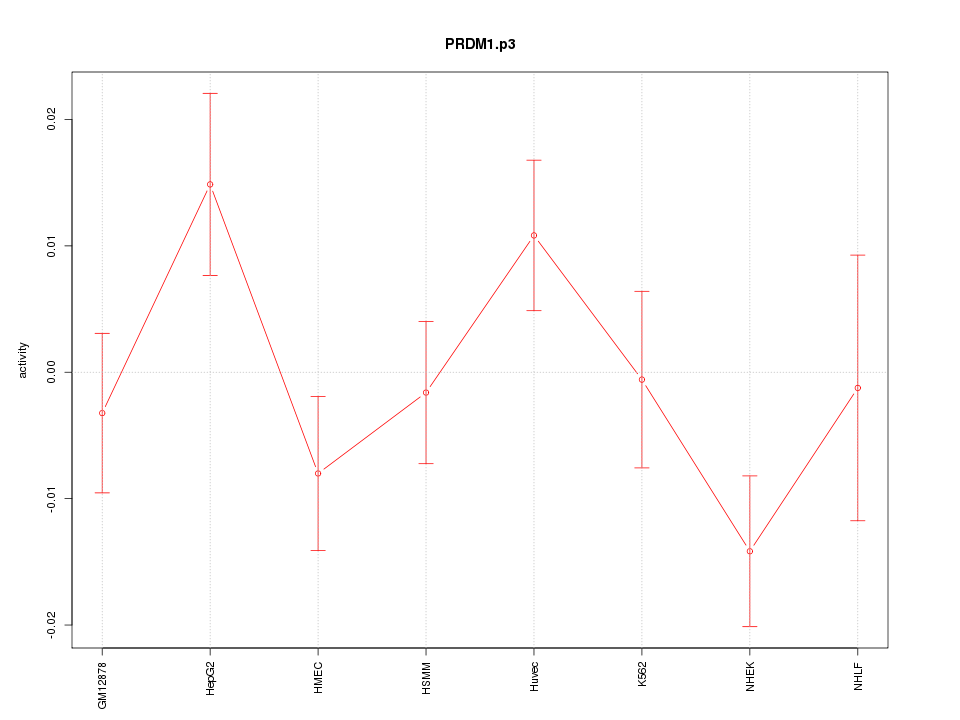
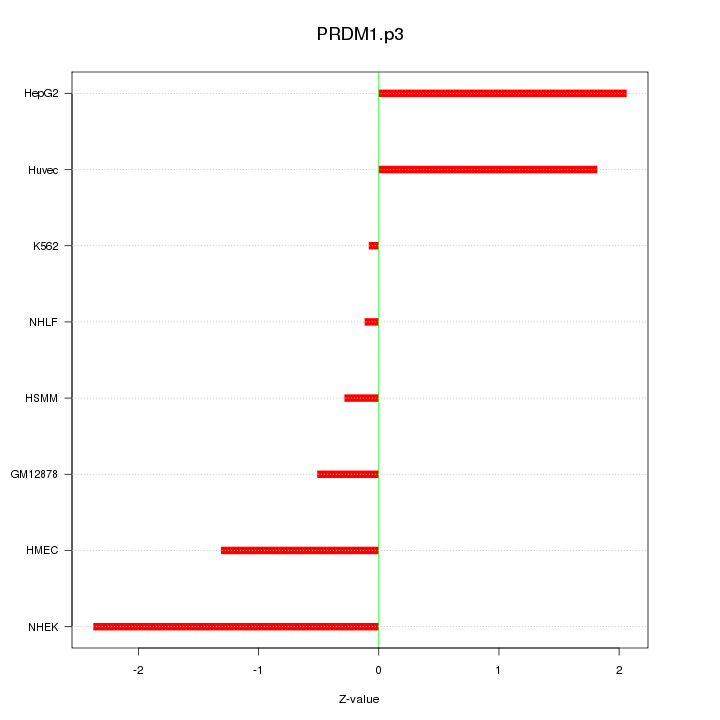
chr2_-_21106291
|
3.005
|
|
APOB
|
apolipoprotein B (including Ag(x) antigen)
|
|
chr5_+_73145098
|
1.583
|
|
RGNEF
|
190 kDa guanine nucleotide exchange factor
|
|
chr15_+_94677572
|
1.568
|
NM_001145157
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr20_+_56904042
|
1.478
|
|
GNAS
|
GNAS complex locus
|
|
chr1_+_210848763
|
1.453
|
|
ATF3
|
activating transcription factor 3
|
|
chr14_+_23675233
|
1.326
|
|
PSME1
|
proteasome (prosome, macropain) activator subunit 1 (PA28 alpha)
|
|
chr14_+_23675206
|
1.299
|
NM_006263
NM_176783
|
PSME1
|
proteasome (prosome, macropain) activator subunit 1 (PA28 alpha)
|
|
chr1_+_210848726
|
1.258
|
|
ATF3
|
activating transcription factor 3
|
|
chr1_+_210848592
|
1.257
|
NM_001040619
NM_001674
|
ATF3
|
activating transcription factor 3
|
|
chr2_+_205118801
|
1.232
|
|
PARD3B
|
par-3 partitioning defective 3 homolog B (C. elegans)
|
|
chr1_-_8406288
|
1.217
|
NM_001042682
|
RERE
|
arginine-glutamic acid dipeptide (RE) repeats
|
|
chr12_-_69289889
|
1.125
|
NM_002837
|
PTPRB
|
protein tyrosine phosphatase, receptor type, B
|
|
chr10_+_92970487
|
1.106
|
|
PCGF5
|
polycomb group ring finger 5
|
|
chr1_-_7998265
|
1.055
|
|
ERRFI1
|
ERBB receptor feedback inhibitor 1
|
|
chr1_+_220884257
|
1.021
|
|
MIA3
|
melanoma inhibitory activity family, member 3
|
|
chr2_+_205118961
|
0.952
|
|
PARD3B
|
par-3 partitioning defective 3 homolog B (C. elegans)
|
|
chr6_+_126282078
|
0.926
|
|
NCOA7
|
nuclear receptor coactivator 7
|
|
chr9_-_34700062
|
0.917
|
NM_002989
|
CCL21
|
chemokine (C-C motif) ligand 21
|
|
chr20_+_13924263
|
0.895
|
|
MACROD2
|
MACRO domain containing 2
|
|
chr1_-_142536082
|
0.814
|
|
|
|
|
chr11_-_77730573
|
0.807
|
NM_012296
|
GAB2
|
GRB2-associated binding protein 2
|
|
chr1_+_166172545
|
0.786
|
|
DCAF6
|
DDB1 and CUL4 associated factor 6
|
|
chr10_+_92970302
|
0.782
|
NM_032373
|
PCGF5
|
polycomb group ring finger 5
|
|
chr4_-_153523113
|
0.781
|
NM_001013415
|
FBXW7
|
F-box and WD repeat domain containing 7
|
|
chr2_+_205118710
|
0.740
|
NM_057177
NM_152526
NM_205863
|
PARD3B
|
par-3 partitioning defective 3 homolog B (C. elegans)
|
|
chr11_+_35641234
|
0.730
|
|
TRIM44
|
tripartite motif containing 44
|
|
chr3_+_186529376
|
0.729
|
|
MAP3K13
|
mitogen-activated protein kinase kinase kinase 13
|
|
chr4_+_146623405
|
0.720
|
NM_001003688
|
SMAD1
|
SMAD family member 1
|
|
chr20_+_56903595
|
0.698
|
|
GNAS
|
GNAS complex locus
|
|
chr1_+_166172420
|
0.689
|
NM_001017977
NM_001198956
NM_001198957
NM_018442
|
DCAF6
|
DDB1 and CUL4 associated factor 6
|
|
chr14_-_23771399
|
0.668
|
|
NEDD8
|
neural precursor cell expressed, developmentally down-regulated 8
|
|
chr14_+_57781308
|
0.645
|
NM_002788
NM_152132
|
PSMA3
|
proteasome (prosome, macropain) subunit, alpha type, 3
|
|
chr1_-_111548440
|
0.643
|
|
DENND2D
|
DENN/MADD domain containing 2D
|
|
chr14_-_23771312
|
0.629
|
|
NEDD8
|
neural precursor cell expressed, developmentally down-regulated 8
|
|
chr2_-_45648608
|
0.623
|
|
SRBD1
|
S1 RNA binding domain 1
|
|
chr8_+_97575196
|
0.604
|
|
SDC2
|
syndecan 2
|
|
chr16_-_46738061
|
0.601
|
NM_033226
|
ABCC12
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 12
|
|
chr3_+_120495906
|
0.583
|
NM_020754
|
ARHGAP31
|
Rho GTPase activating protein 31
|
|
chr15_-_35180656
|
0.579
|
NM_002399
|
MEIS2
|
Meis homeobox 2
|
|
chr2_+_230989114
|
0.567
|
NM_001080391
NM_003113
|
SP100
|
SP100 nuclear antigen
|
|
chr8_+_96215118
|
0.564
|
NM_024613
|
PLEKHF2
|
pleckstrin homology domain containing, family F (with FYVE domain) member 2
|
|
chr7_-_11643113
|
0.562
|
|
THSD7A
|
thrombospondin, type I, domain containing 7A
|
|
chr2_+_230989177
|
0.561
|
|
SP100
|
SP100 nuclear antigen
|
|
chr8_+_97575071
|
0.555
|
|
SDC2
|
syndecan 2
|
|
chr2_+_165858586
|
0.550
|
NM_021007
|
SCN2A
|
sodium channel, voltage-gated, type II, alpha subunit
|
|
chr20_+_13924014
|
0.546
|
NM_080676
|
MACROD2
|
MACRO domain containing 2
|
|
chr14_-_23685629
|
0.546
|
NM_002818
|
PSME2
|
proteasome (prosome, macropain) activator subunit 2 (PA28 beta)
|
|
chr14_+_57781352
|
0.544
|
|
PSMA3
|
proteasome (prosome, macropain) subunit, alpha type, 3
|
|
chr2_+_234209838
|
0.543
|
NM_019075
|
UGT1A10
|
UDP glucuronosyltransferase 1 family, polypeptide A10
|
|
chr14_-_23771401
|
0.541
|
NM_006156
|
NEDD8
|
neural precursor cell expressed, developmentally down-regulated 8
|
|
chr11_+_32069183
|
0.539
|
|
RCN1
|
reticulocalbin 1, EF-hand calcium binding domain
|
|
chr2_-_19421852
|
0.534
|
NM_145260
|
OSR1
|
odd-skipped related 1 (Drosophila)
|
|
chr14_+_23771464
|
0.534
|
NM_001002000
NM_001002001
|
GMPR2
|
guanosine monophosphate reductase 2
|
|
chr20_+_336693
|
0.532
|
NM_006462
NM_031229
|
RBCK1
|
RanBP-type and C3HC4-type zinc finger containing 1
|
|
chr20_+_336963
|
0.527
|
|
RBCK1
|
RanBP-type and C3HC4-type zinc finger containing 1
|
|
chr4_+_76868763
|
0.525
|
|
USO1
|
USO1 vesicle docking protein homolog (yeast)
|
|
chr14_+_57781210
|
0.517
|
|
PSMA3
|
proteasome (prosome, macropain) subunit, alpha type, 3
|
|
chr2_+_230900121
|
0.512
|
NM_138402
|
SP140L
|
SP140 nuclear body protein-like
|
|
chr16_+_49333513
|
0.510
|
NM_001042412
|
CYLD
|
cylindromatosis (turban tumor syndrome)
|
|
chr2_+_230989150
|
0.502
|
|
SP100
|
SP100 nuclear antigen
|
|
chr6_+_32929902
|
0.496
|
NM_002800
|
PSMB9
|
proteasome (prosome, macropain) subunit, beta type, 9 (large multifunctional peptidase 2)
|
|
chr10_-_35143900
|
0.487
|
NM_001184785
NM_001184786
NM_001184787
NM_001184788
NM_001184789
NM_001184790
NM_001184791
NM_001184792
NM_001184793
NM_001184794
NM_019619
|
PARD3
|
par-3 partitioning defective 3 homolog (C. elegans)
|
|
chr8_+_96215144
|
0.474
|
|
PLEKHF2
|
pleckstrin homology domain containing, family F (with FYVE domain) member 2
|
|
chr7_-_42938255
|
0.467
|
|
PSMA2
|
proteasome (prosome, macropain) subunit, alpha type, 2
|
|
chr8_+_97575052
|
0.466
|
NM_002998
|
SDC2
|
syndecan 2
|
|
chr1_-_146398501
|
0.461
|
|
LOC100286793
FLJ39739
|
hypothetical LOC100286793
hypothetical FLJ39739
|
|
chr1_+_149638633
|
0.459
|
NM_002796
|
PSMB4
|
proteasome (prosome, macropain) subunit, beta type, 4
|
|
chr20_+_336717
|
0.456
|
|
RBCK1
|
RanBP-type and C3HC4-type zinc finger containing 1
|
|
chr8_+_97575244
|
0.456
|
|
SDC2
|
syndecan 2
|
|
chr16_+_49333461
|
0.453
|
NM_001042355
NM_015247
|
CYLD
|
cylindromatosis (turban tumor syndrome)
|
|
chr5_+_161209969
|
0.452
|
NM_001127647
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1
|
|
chr7_-_42938304
|
0.447
|
NM_002787
|
PSMA2
|
proteasome (prosome, macropain) subunit, alpha type, 2
|
|
chr15_-_53996597
|
0.444
|
NM_198400
|
NEDD4
|
neural precursor cell expressed, developmentally down-regulated 4
|
|
chr6_-_112682401
|
0.428
|
|
LAMA4
|
laminin, alpha 4
|
|
chr9_-_116608113
|
0.423
|
NM_005118
|
TNFSF15
|
tumor necrosis factor (ligand) superfamily, member 15
|
|
chr6_-_112682459
|
0.423
|
NM_001105206
NM_001105207
NM_001105208
NM_001105209
NM_002290
|
LAMA4
|
laminin, alpha 4
|
|
chr4_+_76868830
|
0.422
|
|
USO1
|
USO1 vesicle docking protein homolog (yeast)
|
|
chr1_+_158063130
|
0.418
|
|
SLAMF8
|
SLAM family member 8
|
|
chr7_+_42938463
|
0.417
|
NM_031903
|
MRPL32
|
mitochondrial ribosomal protein L32
|
|
chr9_+_96806235
|
0.412
|
|
C9orf3
|
chromosome 9 open reading frame 3
|
|
chr9_-_16717887
|
0.410
|
|
BNC2
|
basonuclin 2
|
|
chr8_-_79880095
|
0.400
|
NM_000880
|
IL7
|
interleukin 7
|
|
chr4_+_142777172
|
0.394
|
NM_000585
NM_172174
|
IL15
|
interleukin 15
|
|
chr1_-_143232464
|
0.390
|
|
|
|
|
chr12_+_25096472
|
0.389
|
NM_006152
|
LRMP
|
lymphoid-restricted membrane protein
|
|
chr18_+_18989786
|
0.388
|
NM_138375
|
CABLES1
|
Cdk5 and Abl enzyme substrate 1
|
|
chr6_-_112682573
|
0.386
|
|
LAMA4
|
laminin, alpha 4
|
|
chr11_+_28088406
|
0.377
|
|
METTL15
|
methyltransferase like 15
|
|
chr2_+_6935246
|
0.375
|
NM_080657
|
RSAD2
|
radical S-adenosyl methionine domain containing 2
|
|
chr8_+_39890484
|
0.374
|
NM_002164
|
IDO1
|
indoleamine 2,3-dioxygenase 1
|
|
chr20_+_336941
|
0.372
|
|
RBCK1
|
RanBP-type and C3HC4-type zinc finger containing 1
|
|
chr13_+_107720475
|
0.371
|
|
TNFSF13B
|
tumor necrosis factor (ligand) superfamily, member 13b
|
|
chr5_-_142795269
|
0.360
|
NM_001018074
NM_001018075
NM_001018077
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor)
|
|
chr9_+_108665168
|
0.357
|
NM_021224
|
ZNF462
|
zinc finger protein 462
|
|
chr14_+_23982017
|
0.353
|
|
|
|
|
chr2_+_234191029
|
0.348
|
NM_019076
|
UGT1A8
|
UDP glucuronosyltransferase 1 family, polypeptide A8
|
|
chr1_-_146543016
|
0.346
|
|
|
|
|
chr2_-_189752575
|
0.343
|
NM_000393
|
COL5A2
|
collagen, type V, alpha 2
|
|
chr1_+_158063278
|
0.335
|
|
SLAMF8
|
SLAM family member 8
|
|
chr3_-_161584137
|
0.327
|
NM_001190242
|
IFT80
|
intraflagellar transport 80 homolog (Chlamydomonas)
|
|
chr8_-_145132551
|
0.323
|
|
PARP10
|
poly (ADP-ribose) polymerase family, member 10
|
|
chr8_-_145132582
|
0.316
|
NM_032789
|
PARP10
|
poly (ADP-ribose) polymerase family, member 10
|
|
chr5_-_37220702
|
0.312
|
|
C5orf42
|
chromosome 5 open reading frame 42
|
|
chr1_-_166171838
|
0.307
|
NM_015415
|
BRP44
|
brain protein 44
|
|
chr4_-_76868581
|
0.302
|
|
|
|
|
chr1_-_142535878
|
0.298
|
|
LOC100286793
FLJ39739
|
hypothetical LOC100286793
hypothetical FLJ39739
|
|
chr12_+_68923043
|
0.297
|
NM_014515
|
CNOT2
|
CCR4-NOT transcription complex, subunit 2
|
|
chr20_+_337346
|
0.291
|
|
RBCK1
|
RanBP-type and C3HC4-type zinc finger containing 1
|
|
chr4_+_76868850
|
0.287
|
NM_003715
|
USO1
|
USO1 vesicle docking protein homolog (yeast)
|
|
chr4_+_88075176
|
0.287
|
NM_001166693
|
AFF1
|
AF4/FMR2 family, member 1
|
|
chr14_-_68329407
|
0.285
|
NM_004926
|
ZFP36L1
|
zinc finger protein 36, C3H type-like 1
|
|
chr5_+_139869819
|
0.284
|
|
|
|
|
chr1_-_152847256
|
0.283
|
|
ADAR
|
adenosine deaminase, RNA-specific
|
|
chr11_-_28086277
|
0.281
|
NM_031217
|
KIF18A
|
kinesin family member 18A
|
|
chr14_+_101297887
|
0.280
|
NM_001161725
NM_001161726
|
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma
|
|
chr11_-_40272239
|
0.279
|
NM_020929
|
LRRC4C
|
leucine rich repeat containing 4C
|
|
chr11_-_28086222
|
0.275
|
|
KIF18A
|
kinesin family member 18A
|
|
chr3_+_115099007
|
0.269
|
NM_001172105
|
GRAMD1C
|
GRAM domain containing 1C
|
|
chr14_+_72594985
|
0.268
|
|
RBM25
|
RNA binding motif protein 25
|
|
chr11_-_28086263
|
0.268
|
|
KIF18A
|
kinesin family member 18A
|
|
chr11_+_64946848
|
0.268
|
|
|
|
|
chr21_+_42807052
|
0.267
|
|
SLC37A1
|
solute carrier family 37 (glycerol-3-phosphate transporter), member 1
|
|
chr6_-_112682038
|
0.266
|
|
LAMA4
|
laminin, alpha 4
|
|
chr1_-_166171702
|
0.265
|
|
BRP44
|
brain protein 44
|
|
chr3_+_189354167
|
0.264
|
NM_001167672
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr16_-_71650034
|
0.258
|
NM_001164766
|
ZFHX3
|
zinc finger homeobox 3
|
|
chr16_+_55580910
|
0.255
|
NM_032206
|
NLRC5
|
NLR family, CARD domain containing 5
|
|
chr10_-_6059460
|
0.249
|
NM_002189
NM_172200
|
IL15RA
|
interleukin 15 receptor, alpha
|
|
chr13_+_107720206
|
0.241
|
|
TNFSF13B
|
tumor necrosis factor (ligand) superfamily, member 13b
|
|
chr19_-_53239030
|
0.236
|
NM_019855
|
CABP5
|
calcium binding protein 5
|
|
chr8_-_29176490
|
0.233
|
NM_015254
|
KIF13B
|
kinesin family member 13B
|
|
chr11_+_64946820
|
0.227
|
|
NEAT1
|
nuclear paraspeckle assembly transcript 1 (non-protein coding)
|
|
chr10_+_63478974
|
0.227
|
|
ARID5B
|
AT rich interactive domain 5B (MRF1-like)
|
|
chr1_-_152847301
|
0.226
|
NM_001111
NM_015840
NM_015841
|
ADAR
|
adenosine deaminase, RNA-specific
|
|
chr14_+_72595020
|
0.225
|
|
RBM25
|
RNA binding motif protein 25
|
|
chr11_-_5662762
|
0.225
|
NM_033034
NM_033092
NM_033093
|
TRIM5
|
tripartite motif containing 5
|
|
chr1_-_143232526
|
0.223
|
|
FAM91A2
|
family with sequence similarity 91, member A2
|
|
chr10_+_69539255
|
0.217
|
NM_032578
|
MYPN
|
myopalladin
|
|
chr3_-_189354510
|
0.217
|
|
LOC339929
|
hypothetical LOC339929
|
|
chr12_+_11694172
|
0.216
|
|
ETV6
|
ets variant 6
|
|
chr10_+_91051685
|
0.213
|
NM_001547
|
IFIT2
|
interferon-induced protein with tetratricopeptide repeats 2
|
|
chr6_-_11887265
|
0.212
|
NM_001143948
NM_032744
|
C6orf105
|
chromosome 6 open reading frame 105
|
|
chr6_-_33389825
|
0.208
|
|
TAPBP
|
TAP binding protein (tapasin)
|
|
chr11_-_4371463
|
0.208
|
NM_003141
|
TRIM21
|
tripartite motif containing 21
|
|
chr1_-_143232275
|
0.204
|
|
LOC728855
|
hypothetical LOC728855
|
|
chr11_+_28086373
|
0.203
|
NM_001113528
NM_152636
|
METTL15
|
methyltransferase like 15
|
|
chr16_-_70763849
|
0.201
|
NM_001160213
|
PMFBP1
|
polyamine modulated factor 1 binding protein 1
|
|
chr3_-_11598829
|
0.197
|
NM_001128220
|
VGLL4
|
vestigial like 4 (Drosophila)
|
|
chr1_+_158063102
|
0.196
|
NM_020125
|
SLAMF8
|
SLAM family member 8
|
|
chr2_-_202216451
|
0.194
|
NM_001044385
|
ALS2CR4
|
amyotrophic lateral sclerosis 2 (juvenile) chromosome region, candidate 4
|
|
chr6_-_28663045
|
0.187
|
NM_052923
|
SCAND3
|
SCAN domain containing 3
|
|
chr3_+_108724472
|
0.187
|
NM_001142568
NM_020235
|
BBX
|
bobby sox homolog (Drosophila)
|
|
chr9_+_101717278
|
0.186
|
|
STX17
|
syntaxin 17
|
|
chr12_-_55134692
|
0.174
|
NM_012064
|
MIP
|
major intrinsic protein of lens fiber
|
|
chr5_+_49998528
|
0.173
|
NM_001178056
NM_024615
|
PARP8
|
poly (ADP-ribose) polymerase family, member 8
|
|
chr6_-_33389768
|
0.171
|
|
TAPBP
|
TAP binding protein (tapasin)
|
|
chr1_-_159435440
|
0.168
|
NM_005099
|
ADAMTS4
|
ADAM metallopeptidase with thrombospondin type 1 motif, 4
|
|
chr12_-_55791443
|
0.163
|
NM_001178078
NM_001178080
NM_001178081
NM_003153
|
STAT6
|
signal transducer and activator of transcription 6, interleukin-4 induced
|
|
chr11_-_85015961
|
0.161
|
NM_001142699
|
DLG2
|
discs, large homolog 2 (Drosophila)
|
|
chr6_-_105691675
|
0.160
|
|
BVES
|
blood vessel epicardial substance
|
|
chr13_+_107719977
|
0.160
|
NM_001145645
NM_006573
|
TNFSF13B
|
tumor necrosis factor (ligand) superfamily, member 13b
|
|
chr2_-_37164919
|
0.159
|
|
HEATR5B
|
HEAT repeat containing 5B
|
|
chr3_+_69998064
|
0.158
|
NM_198177
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr11_-_125153914
|
0.158
|
NM_212555
|
PATE2
|
prostate and testis expressed 2
|
|
chr14_-_22549168
|
0.158
|
NM_001130706
NM_001130708
NM_021944
|
C14orf93
|
chromosome 14 open reading frame 93
|
|
chr2_-_144711375
|
0.157
|
NM_024659
|
GTDC1
|
glycosyltransferase-like domain containing 1
|
|
chr14_-_23981493
|
0.152
|
NM_020195
|
SDR39U1
|
short chain dehydrogenase/reductase family 39U, member 1
|
|
chr7_+_134483305
|
0.152
|
NM_018295
|
TMEM140
|
transmembrane protein 140
|
|
chr7_-_105106844
|
0.152
|
NM_138495
|
ATXN7L1
|
ataxin 7-like 1
|
|
chr9_+_109085372
|
0.151
|
|
RAD23B
|
RAD23 homolog B (S. cerevisiae)
|
|
chr9_-_14858974
|
0.151
|
|
FREM1
|
FRAS1 related extracellular matrix 1
|
|
chr16_-_10696285
|
0.150
|
NM_144674
|
TEKT5
|
tektin 5
|
|
chr1_-_116185215
|
0.145
|
NM_005599
|
NHLH2
|
nescient helix loop helix 2
|
|
chr22_+_38903825
|
0.143
|
NM_001162501
NM_015088
|
TNRC6B
|
trinucleotide repeat containing 6B
|
|
chr16_-_70763524
|
0.140
|
NM_031293
|
PMFBP1
|
polyamine modulated factor 1 binding protein 1
|
|
chr7_+_89681069
|
0.140
|
NM_001040666
|
STEAP2
|
six transmembrane epithelial antigen of the prostate 2
|
|
chr12_+_27511009
|
0.137
|
NM_001145010
|
C12orf70
|
chromosome 12 open reading frame 70
|
|
chr11_+_5602782
|
0.135
|
NM_021616
|
TRIM34
|
tripartite motif containing 34
|
|
chr8_-_9046460
|
0.134
|
|
PPP1R3B
|
protein phosphatase 1, regulatory (inhibitor) subunit 3B
|
|
chr1_-_146543102
|
0.133
|
|
|
|
|
chr1_+_159435728
|
0.131
|
NM_004550
|
NDUFS2
|
NADH dehydrogenase (ubiquinone) Fe-S protein 2, 49kDa (NADH-coenzyme Q reductase)
|
|
chr15_-_40537002
|
0.131
|
NM_022473
|
ZFP106
|
zinc finger protein 106 homolog (mouse)
|
|
chr17_+_53602013
|
0.130
|
NM_001004707
|
OR4D2
|
olfactory receptor, family 4, subfamily D, member 2
|
|
chr7_+_134114701
|
0.129
|
NM_004342
NM_033138
NM_033157
|
CALD1
|
caldesmon 1
|
|
chr12_-_55791388
|
0.128
|
|
STAT6
|
signal transducer and activator of transcription 6, interleukin-4 induced
|
|
chr8_+_143805595
|
0.128
|
NM_016647
|
C8orf55
|
chromosome 8 open reading frame 55
|
|
chr10_-_113923507
|
0.127
|
|
GPAM
|
glycerol-3-phosphate acyltransferase, mitochondrial
|
|
chr9_+_109085337
|
0.127
|
NM_002874
|
RAD23B
|
RAD23 homolog B (S. cerevisiae)
|
|
chr20_-_47327998
|
0.126
|
NM_021035
|
ZNFX1
|
zinc finger, NFX1-type containing 1
|
|
chr8_+_97726757
|
0.123
|
|
PGCP
|
plasma glutamate carboxypeptidase
|
|
chr19_+_61202902
|
0.123
|
NM_153447
|
NLRP5
|
NLR family, pyrin domain containing 5
|
|
chr14_+_72594940
|
0.123
|
NM_021239
|
RBM25
|
RNA binding motif protein 25
|
|
chr7_-_158304062
|
0.120
|
|
ESYT2
|
extended synaptotagmin-like protein 2
|
|
chr1_-_242073094
|
0.119
|
NM_005465
NM_181690
|
AKT3
|
v-akt murine thymoma viral oncogene homolog 3 (protein kinase B, gamma)
|
|
chr20_-_34102250
|
0.119
|
|
LOC647979
|
hypothetical LOC647979
|
|
chr6_+_118976134
|
0.119
|
NM_002667
|
PLN
|
phospholamban
|
|
chr2_+_17585462
|
0.118
|
|
VSNL1
|
visinin-like 1
|
|
chr7_-_86686754
|
0.118
|
NM_024315
|
C7orf23
|
chromosome 7 open reading frame 23
|
|
chr8_-_143805371
|
0.117
|
|
LOC100288181
|
hypothetical LOC100288181
|
|
chr6_-_32251877
|
0.116
|
NM_006411
|
AGPAT1
|
1-acylglycerol-3-phosphate O-acyltransferase 1 (lysophosphatidic acid acyltransferase, alpha)
|