|
chr11_-_16380967
|
3.627
|
NM_001145819
NM_017508
|
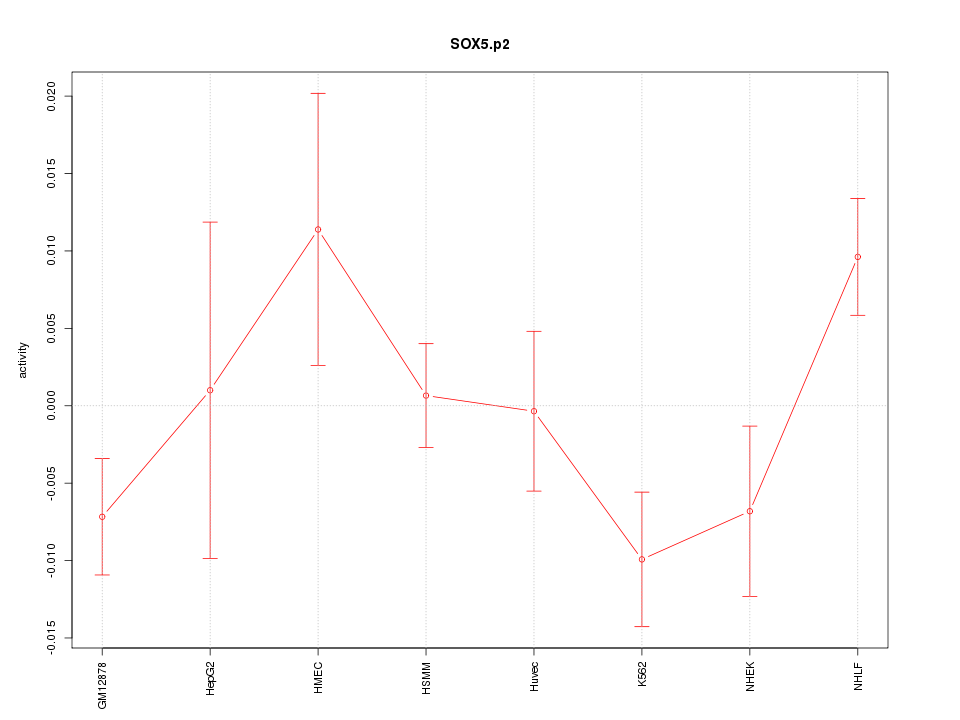
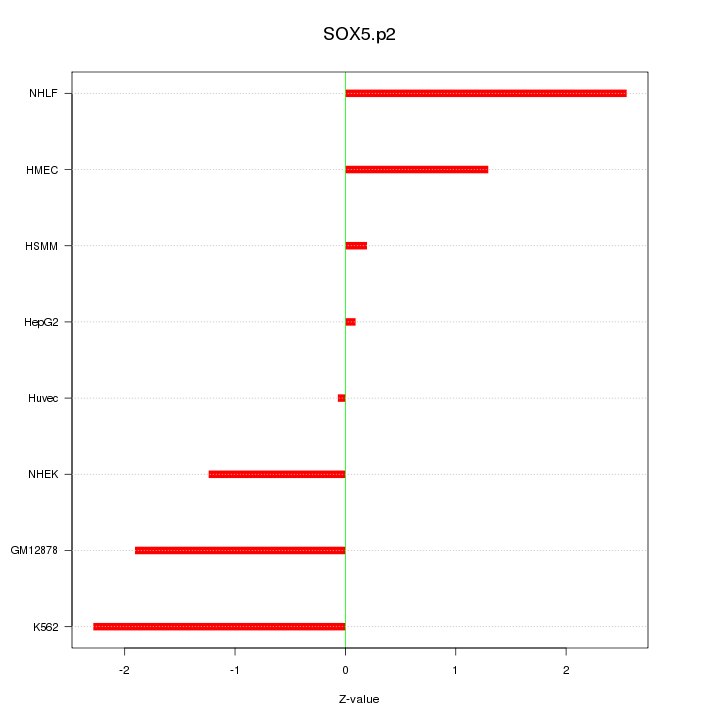
SOX6
|
SRY (sex determining region Y)-box 6
|
|
chr7_-_11838260
|
3.244
|
NM_015204
|
THSD7A
|
thrombospondin, type I, domain containing 7A
|
|
chr4_+_42590038
|
3.005
|
NM_001080476
|
GRXCR1
|
glutaredoxin, cysteine rich 1
|
|
chr3_+_174598937
|
2.124
|
NM_014932
|
NLGN1
|
neuroligin 1
|
|
chr4_-_101658094
|
2.099
|
NM_001159694
NM_016242
|
EMCN
|
endomucin
|
|
chr3_+_88270951
|
1.944
|
NM_018293
|
ZNF654
|
zinc finger protein 654
|
|
chr11_-_51269023
|
1.924
|
NM_001005272
|
OR4A5
|
olfactory receptor, family 4, subfamily A, member 5
|
|
chr2_+_166137091
|
1.897
|
NM_024969
|
CSRNP3
|
cysteine-serine-rich nuclear protein 3
|
|
chr4_-_105635499
|
1.806
|
|
CXXC4
|
CXXC finger protein 4
|
|
chr12_-_11399776
|
1.805
|
NM_005039
NM_199353
NM_199354
|
PRB1
|
proline-rich protein BstNI subfamily 1
|
|
chr15_+_47502666
|
1.783
|
NM_002009
|
FGF7
|
fibroblast growth factor 7
|
|
chr3_-_121065112
|
1.778
|
|
|
|
|
chr10_+_96433240
|
1.713
|
NM_000772
NM_001128925
|
CYP2C18
|
cytochrome P450, family 2, subfamily C, polypeptide 18
|
|
chrX_-_151370325
|
1.709
|
NM_000808
|
GABRA3
|
gamma-aminobutyric acid (GABA) A receptor, alpha 3
|
|
chr2_-_225073357
|
1.646
|
|
CUL3
|
cullin 3
|
|
chr1_-_51660328
|
1.616
|
NM_001159969
|
EPS15
|
epidermal growth factor receptor pathway substrate 15
|
|
chr20_-_21326046
|
1.614
|
NM_033176
|
NKX2-4
|
NK2 homeobox 4
|
|
chr11_-_121492010
|
1.602
|
NM_001001786
|
BLID
|
BH3-like motif containing, cell death inducer
|
|
chr9_-_85773899
|
1.584
|
|
HNRNPK
|
heterogeneous nuclear ribonucleoprotein K
|
|
chr10_+_49184658
|
1.576
|
|
MAPK8
|
mitogen-activated protein kinase 8
|
|
chr11_-_48330574
|
1.571
|
NM_001005513
|
OR4C45
|
olfactory receptor, family 4, subfamily C, member 45
|
|
chr11_+_101488380
|
1.557
|
NM_001195045
|
YAP1
|
Yes-associated protein 1
|
|
chr4_+_71096830
|
1.544
|
NM_017855
|
ODAM
|
odontogenic, ameloblast asssociated
|
|
chr12_-_11354620
|
1.527
|
NM_002723
|
PRB4
|
proline-rich protein BstNI subfamily 4
|
|
chr18_-_29412148
|
1.512
|
|
|
|
|
chr11_-_7684516
|
1.501
|
NM_198185
|
OVCH2
|
ovochymase 2 (gene/pseudogene)
|
|
chr1_+_198975227
|
1.497
|
NM_203459
|
CAMSAP1L1
|
calmodulin regulated spectrin-associated protein 1-like 1
|
|
chr2_-_55053398
|
1.446
|
|
|
|
|
chr15_+_22751162
|
1.412
|
NM_003097
NM_022804
NM_005678
|
SNRPN
SNURF
|
small nuclear ribonucleoprotein polypeptide N
SNRPN upstream reading frame
|
|
chr14_-_105862080
|
1.383
|
|
IGHV4-31
IGHA1
IGHD
IGHG1
IGHG2
IGHG3
IGHM
SCFV
|
immunoglobulin heavy variable 4-31
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant delta
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant gamma 2 (G2m marker)
immunoglobulin heavy constant gamma 3 (G3m marker)
immunoglobulin heavy constant mu
single-chain Fv fragment
|
|
chr4_+_110968594
|
1.365
|
NM_006583
|
RRH
|
retinal pigment epithelium-derived rhodopsin homolog
|
|
chr8_+_104900591
|
1.354
|
NM_014677
|
RIMS2
|
regulating synaptic membrane exocytosis 2
|
|
chr3_+_89239494
|
1.354
|
|
EPHA3
|
EPH receptor A3
|
|
chr10_+_104525877
|
1.349
|
NM_017787
|
C10orf26
|
chromosome 10 open reading frame 26
|
|
chr1_-_150564302
|
1.334
|
NM_002016
|
FLG
|
filaggrin
|
|
chr9_-_21375258
|
1.330
|
NM_000605
|
IFNA2
|
interferon, alpha 2
|
|
chr17_-_44058612
|
1.283
|
NM_024017
|
HOXB9
|
homeobox B9
|
|
chr17_+_3065661
|
1.274
|
NM_014565
|
OR1A1
|
olfactory receptor, family 1, subfamily A, member 1
|
|
chr15_-_23204887
|
1.249
|
NM_000462
|
UBE3A
|
ubiquitin protein ligase E3A
|
|
chr9_+_108665168
|
1.248
|
NM_021224
|
ZNF462
|
zinc finger protein 462
|
|
chr11_-_55460451
|
1.206
|
NM_006637
|
OR5I1
|
olfactory receptor, family 5, subfamily I, member 1
|
|
chr9_+_102244005
|
1.201
|
NM_001198812
|
C9orf3-TMEFF1
C9orf30
|
C9orf3-TMEFF1 readthrough
chromosome 9 open reading frame 30
|
|
chr21_-_30583702
|
1.199
|
NM_001128598
|
KRTAP25-1
|
keratin associated protein 25-1
|
|
chr3_+_89239363
|
1.198
|
NM_005233
NM_182644
|
EPHA3
|
EPH receptor A3
|
|
chr5_-_111340421
|
1.188
|
NM_001142474
NM_001142475
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr1_-_7998265
|
1.173
|
|
ERRFI1
|
ERBB receptor feedback inhibitor 1
|
|
chr10_-_98311813
|
1.172
|
|
TM9SF3
|
transmembrane 9 superfamily member 3
|
|
chr2_-_63518589
|
1.166
|
NM_001042692
|
WDPCP
|
WD repeat containing planar cell polarity effector
|
|
chrX_+_55951280
|
1.164
|
|
|
|
|
chr12_-_23995109
|
1.162
|
|
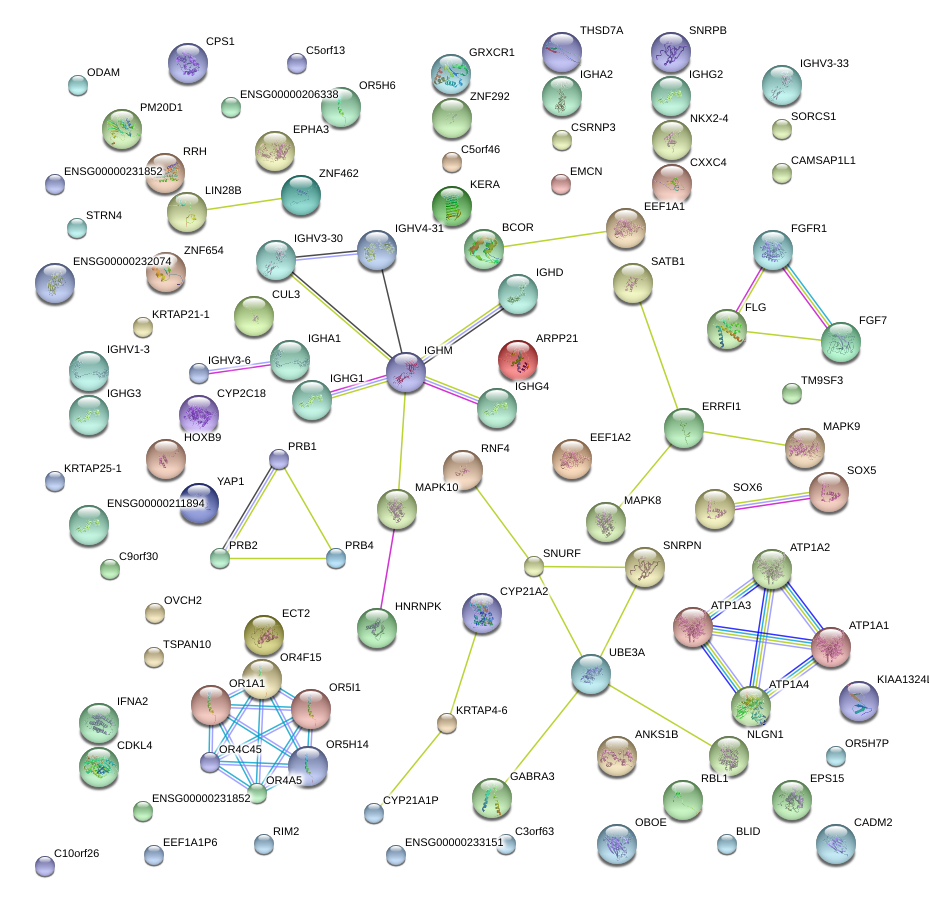
SOX5
|
SRY (sex determining region Y)-box 5
|
|
chr6_+_105511615
|
1.157
|
NM_001004317
|
LIN28B
|
lin-28 homolog B (C. elegans)
|
|
chr12_-_23995220
|
1.153
|
|
SOX5
|
SRY (sex determining region Y)-box 5
|
|
chr10_-_98280299
|
1.141
|
|
TM9SF3
|
transmembrane 9 superfamily member 3
|
|
chr7_-_86526880
|
1.136
|
NM_001142749
|
KIAA1324L
|
KIAA1324-like
|
|
chr2_-_39310176
|
1.093
|
NM_001009565
|
CDKL4
|
cyclin-dependent kinase-like 4
|
|
chr12_-_89976261
|
1.093
|
NM_007035
|
KERA
|
keratocan
|
|
chr5_-_147266257
|
1.090
|
NM_206966
|
C5orf46
|
chromosome 5 open reading frame 46
|
|
chr3_+_99350859
|
1.082
|
NM_001005514
|
OR5H14
|
olfactory receptor, family 5, subfamily H, member 14
|
|
chr7_-_22518200
|
1.081
|
|
EEF1A1
|
eukaryotic translation elongation factor 1 alpha 1
|
|
chr6_+_88026338
|
1.081
|
|
ZNF292
|
zinc finger protein 292
|
|
chr3_-_18455207
|
1.074
|
NM_001131010
|
SATB1
|
SATB homeobox 1
|
|
chr12_-_97812633
|
1.073
|
|
ANKS1B
|
ankyrin repeat and sterile alpha motif domain containing 1B
|
|
chr10_-_108914277
|
1.044
|
NM_001013031
NM_052918
|
SORCS1
|
sortilin-related VPS10 domain containing receptor 1
|
|
chr3_-_56673113
|
1.042
|
NM_015224
|
C3orf63
|
chromosome 3 open reading frame 63
|
|
chr3_+_85858321
|
1.041
|
NM_153184
|
CADM2
|
cell adhesion molecule 2
|
|
chr1_+_116727514
|
1.040
|
NM_001160234
|
ATP1A1
|
ATPase, Na+/K+ transporting, alpha 1 polypeptide
|
|
chrX_-_39921525
|
1.035
|
NM_001123383
NM_001123384
|
BCOR
|
BCL6 corepressor
|
|
chr15_+_100175888
|
1.030
|
NM_001001674
|
OR4F15
|
olfactory receptor, family 4, subfamily F, member 15
|
|
chr3_+_173954940
|
1.020
|
NM_018098
|
ECT2
|
epithelial cell transforming sequence 2 oncogene
|
|
chr21_-_31049566
|
1.020
|
NM_181619
|
KRTAP21-1
|
keratin associated protein 21-1
|
|
chr2_+_211166323
|
1.018
|
NM_001122634
|
CPS1
|
carbamoyl-phosphate synthase 1, mitochondrial
|
|
chr14_-_106184988
|
1.016
|
|
IGHA1
IGHG1
|
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr9_-_28709302
|
1.012
|
NM_152570
|
LINGO2
|
leucine rich repeat and Ig domain containing 2
|
|
chr12_+_68028397
|
1.007
|
NM_000239
|
LYZ
|
lysozyme
|
|
chr1_+_145479805
|
1.000
|
NM_004326
|
BCL9
|
B-cell CLL/lymphoma 9
|
|
chr16_+_65938753
|
0.994
|
NM_001161575
|
LRRC36
|
leucine rich repeat containing 36
|
|
chr6_-_46155937
|
0.992
|
NM_001114086
|
CLIC5
|
chloride intracellular channel 5
|
|
chr4_-_113656762
|
0.980
|
NM_024019
|
NEUROG2
|
neurogenin 2
|
|
chr14_-_54948290
|
0.980
|
NM_014924
|
ATG14
|
ATG14 autophagy related 14 homolog (S. cerevisiae)
|
|
chr6_+_154373323
|
0.974
|
NM_001145279
NM_001145280
NM_001145281
|
OPRM1
|
opioid receptor, mu 1
|
|
chr1_-_150353179
|
0.972
|
NM_007113
|
TCHH
|
trichohyalin
|
|
chr2_-_55090759
|
0.970
|
NM_007008
|
RTN4
|
reticulon 4
|
|
chr1_-_49014993
|
0.967
|
NM_024603
|
BEND5
|
BEN domain containing 5
|
|
chr2_+_167468220
|
0.958
|
NM_001079810
NM_152381
|
XIRP2
|
xin actin-binding repeat containing 2
|
|
chr1_-_214963279
|
0.957
|
NM_001438
|
ESRRG
|
estrogen-related receptor gamma
|
|
chr8_-_125785611
|
0.948
|
|
MTSS1
|
metastasis suppressor 1
|
|
chr6_-_128263868
|
0.948
|
NM_001010923
NM_001164685
|
THEMIS
|
thymocyte selection associated
|
|
chr10_-_62002672
|
0.922
|
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr12_+_26017954
|
0.921
|
NM_001164747
|
RASSF8
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8
|
|
chr1_+_183281173
|
0.920
|
NM_007212
|
RNF2
|
ring finger protein 2
|
|
chr14_-_22974707
|
0.916
|
NM_000257
|
MYH7
|
myosin, heavy chain 7, cardiac muscle, beta
|
|
chr3_-_113701073
|
0.903
|
NM_001085357
NM_181780
|
BTLA
|
B and T lymphocyte associated
|
|
chr20_+_20296744
|
0.897
|
NM_002196
|
INSM1
|
insulinoma-associated 1
|
|
chr11_+_5925152
|
0.894
|
NM_001003443
|
OR56A3
|
olfactory receptor, family 56, subfamily A, member 3
|
|
chr5_+_147754467
|
0.893
|
|
FBXO38
|
F-box protein 38
|
|
chr11_+_54891935
|
0.891
|
NM_001005275
|
OR4A15
|
olfactory receptor, family 4, subfamily A, member 15
|
|
chr20_+_23293020
|
0.889
|
NM_022482
|
GZF1
|
GDNF-inducible zinc finger protein 1
|
|
chr10_+_24537725
|
0.889
|
NM_001098501
NM_019590
|
KIAA1217
|
KIAA1217
|
|
chr17_-_36451189
|
0.885
|
NM_030967
|
KRTAP1-3
KRTAP1-1
|
keratin associated protein 1-3
keratin associated protein 1-1
|
|
chr11_+_55799604
|
0.884
|
NM_001004745
|
OR5T1
|
olfactory receptor, family 5, subfamily T, member 1
|
|
chr2_+_62787534
|
0.878
|
NM_001142614
|
EHBP1
|
EH domain binding protein 1
|
|
chr4_+_114190233
|
0.877
|
NM_001148
NM_020977
|
ANK2
|
ankyrin 2, neuronal
|
|
chr11_+_33236435
|
0.874
|
|
HIPK3
|
homeodomain interacting protein kinase 3
|
|
chrX_-_83644054
|
0.871
|
NM_001177478
NM_001177479
NM_144657
|
HDX
|
highly divergent homeobox
|
|
chr12_+_68028407
|
0.859
|
|
LYZ
|
lysozyme
|
|
chr20_+_71230
|
0.857
|
NM_030931
|
DEFB126
|
defensin, beta 126
|
|
chr16_+_68284252
|
0.854
|
|
NFAT5
|
nuclear factor of activated T-cells 5, tonicity-responsive
|
|
chr12_-_11066436
|
0.854
|
NM_176888
|
TAS2R19
|
taste receptor, type 2, member 19
|
|
chr11_+_55842358
|
0.852
|
NM_001005202
|
OR8K3
|
olfactory receptor, family 8, subfamily K, member 3
|
|
chr11_-_123353607
|
0.848
|
NM_001004474
|
OR10S1
|
olfactory receptor, family 10, subfamily S, member 1
|
|
chr16_+_55373241
|
0.845
|
|
NUP93
|
nucleoporin 93kDa
|
|
chr4_-_20594645
|
0.844
|
|
KCNIP4
|
Kv channel interacting protein 4
|
|
chr3_-_130373241
|
0.840
|
|
CNBP
|
CCHC-type zinc finger, nucleic acid binding protein
|
|
chr7_+_93373755
|
0.839
|
NM_021955
|
GNGT1
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1
|
|
chr3_+_150028130
|
0.837
|
NM_001871
|
CPB1
|
carboxypeptidase B1 (tissue)
|
|
chr8_+_103633023
|
0.833
|
NM_024410
|
ODF1
|
outer dense fiber of sperm tails 1
|
|
chr10_-_98263434
|
0.829
|
NM_012465
|
TLL2
|
tolloid-like 2
|
|
chr8_-_72437020
|
0.823
|
NM_000503
|
EYA1
|
eyes absent homolog 1 (Drosophila)
|
|
chr11_+_5862076
|
0.823
|
NM_001005165
|
OR52E4
|
olfactory receptor, family 52, subfamily E, member 4
|
|
chr10_+_97749837
|
0.822
|
NM_001001732
NM_001159747
|
CC2D2B
|
coiled-coil and C2 domain containing 2B
|
|
chr20_+_2892918
|
0.819
|
|
PTPRA
|
protein tyrosine phosphatase, receptor type, A
|
|
chr4_-_83566045
|
0.808
|
|
|
|
|
chr3_-_69212192
|
0.799
|
NM_003968
NM_198195
|
UBA3
|
ubiquitin-like modifier activating enzyme 3
|
|
chr8_-_93144366
|
0.798
|
NM_004349
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr5_-_178977441
|
0.795
|
|
HNRNPH1
|
heterogeneous nuclear ribonucleoprotein H1 (H)
|
|
chr22_-_39189369
|
0.794
|
|
MKL1
|
megakaryoblastic leukemia (translocation) 1
|
|
chr2_-_128138435
|
0.784
|
NM_017980
|
LIMS2
|
LIM and senescent cell antigen-like domains 2
|
|
chr10_-_56230953
|
0.773
|
NM_001142763
NM_001142764
NM_001142765
NM_001142766
NM_001142767
NM_001142768
NM_001142769
NM_001142770
NM_001142771
NM_001142772
NM_001142773
NM_033056
|
PCDH15
|
protocadherin-related 15
|
|
chr3_-_62835601
|
0.771
|
|
|
|
|
chr1_+_81544432
|
0.770
|
|
LPHN2
|
latrophilin 2
|
|
chr9_-_72673753
|
0.770
|
NM_001007470
NM_020952
NM_024971
NM_206944
NM_206945
NM_206946
NM_206947
NM_206948
|
TRPM3
|
transient receptor potential cation channel, subfamily M, member 3
|
|
chr12_-_66905802
|
0.762
|
NM_018402
|
IL26
|
interleukin 26
|
|
chr8_+_74369647
|
0.761
|
NM_172037
|
RDH10
|
retinol dehydrogenase 10 (all-trans)
|
|
chr12_-_11041740
|
0.760
|
NM_176889
|
TAS2R20
|
taste receptor, type 2, member 20
|
|
chr15_+_71953020
|
0.751
|
NM_153356
|
TBC1D21
|
TBC1 domain family, member 21
|
|
chr11_+_22652957
|
0.749
|
|
GAS2
|
growth arrest-specific 2
|
|
chr10_-_74863303
|
0.748
|
NM_001024593
|
ZMYND17
|
zinc finger, MYND-type containing 17
|
|
chr11_-_92570626
|
0.745
|
|
|
|
|
chr3_-_47909001
|
0.741
|
|
MAP4
|
microtubule-associated protein 4
|
|
chr10_+_86121526
|
0.741
|
|
FAM190B
|
family with sequence similarity 190, member B
|
|
chr1_-_246804680
|
0.741
|
NM_001001821
|
OR2T34
|
olfactory receptor, family 2, subfamily T, member 34
|
|
chr2_+_228386800
|
0.740
|
NM_001130046
NM_004591
|
CCL20
|
chemokine (C-C motif) ligand 20
|
|
chr3_+_162697289
|
0.740
|
NM_001080440
|
OTOL1
|
otolin 1 homolog (zebrafish)
|
|
chr2_-_208697441
|
0.738
|
NM_006891
|
CRYGD
|
crystallin, gamma D
|
|
chr4_+_53975597
|
0.737
|
|
FIP1L1
|
FIP1 like 1 (S. cerevisiae)
|
|
chr7_-_82630063
|
0.735
|
NM_014510
NM_033026
|
PCLO
|
piccolo (presynaptic cytomatrix protein)
|
|
chr6_+_89847183
|
0.734
|
|
PNRC1
|
proline-rich nuclear receptor coactivator 1
|
|
chr6_-_128883125
|
0.733
|
NM_002844
NM_001135648
|
PTPRK
|
protein tyrosine phosphatase, receptor type, K
|
|
chr12_-_18134319
|
0.732
|
NM_024730
|
RERGL
|
RERG/RAS-like
|
|
chr5_+_102493155
|
0.731
|
NM_015216
|
PPIP5K2
|
diphosphoinositol pentakisphosphate kinase 2
|
|
chr2_-_164300758
|
0.729
|
NM_018086
|
FIGN
|
fidgetin
|
|
chr10_+_118295417
|
0.727
|
NM_000936
|
PNLIP
|
pancreatic lipase
|
|
chr11_+_36274300
|
0.723
|
NM_001160167
|
PRR5L
|
proline rich 5 like
|
|
chr17_-_3248453
|
0.721
|
NM_003553
|
OR1E1
|
olfactory receptor, family 1, subfamily E, member 1
|
|
chr5_-_146441184
|
0.721
|
NM_181677
NM_181678
|
PPP2R2B
|
protein phosphatase 2, regulatory subunit B, beta
|
|
chr2_+_191009265
|
0.720
|
|
MFSD6
|
major facilitator superfamily domain containing 6
|
|
chr14_-_94669547
|
0.715
|
NM_001195573
|
DICER1
|
dicer 1, ribonuclease type III
|
|
chr15_+_37660568
|
0.712
|
NM_003246
|
THBS1
|
thrombospondin 1
|
|
chr8_+_87947788
|
0.712
|
NM_173538
|
CNBD1
|
cyclic nucleotide binding domain containing 1
|
|
chr14_-_91483757
|
0.711
|
NM_006329
|
FBLN5
|
fibulin 5
|
|
chrX_-_32340291
|
0.705
|
NM_004011
NM_004012
|
DMD
|
dystrophin
|
|
chr15_+_36331411
|
0.705
|
|
SPRED1
|
sprouty-related, EVH1 domain containing 1
|
|
chr10_+_129237602
|
0.702
|
NM_001030013
|
NPS
|
neuropeptide S
|
|
chr11_-_27637771
|
0.695
|
NM_001143816
NM_170735
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr14_+_19414230
|
0.694
|
NM_001005501
|
OR4K2
|
olfactory receptor, family 4, subfamily K, member 2
|
|
chr12_+_21417068
|
0.688
|
NM_000415
|
IAPP
|
islet amyloid polypeptide
|
|
chr18_-_55136673
|
0.686
|
NM_181654
|
CPLX4
|
complexin 4
|
|
chr3_-_66634031
|
0.683
|
|
LRIG1
|
leucine-rich repeats and immunoglobulin-like domains 1
|
|
chrX_-_111970662
|
0.681
|
NM_133265
|
AMOT
|
angiomotin
|
|
chr18_+_29412538
|
0.680
|
NM_030632
|
ASXL3
|
additional sex combs like 3 (Drosophila)
|
|
chr12_-_85756766
|
0.679
|
NM_013244
|
MGAT4C
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative)
|
|
chr13_+_81162030
|
0.679
|
|
|
|
|
chr16_+_21518633
|
0.678
|
|
METTL9
|
methyltransferase like 9
|
|
chr14_-_70345485
|
0.678
|
NM_033141
|
MAP3K9
|
mitogen-activated protein kinase kinase kinase 9
|
|
chr1_+_148603769
|
0.677
|
|
RPRD2
|
regulation of nuclear pre-mRNA domain containing 2
|
|
chr12_+_54132264
|
0.677
|
NM_054105
|
OR6C2
|
olfactory receptor, family 6, subfamily C, member 2
|
|
chr9_-_4142182
|
0.676
|
NM_152629
|
GLIS3
|
GLIS family zinc finger 3
|
|
chr2_+_108230082
|
0.674
|
NM_001008743
|
SULT1C3
|
sulfotransferase family, cytosolic, 1C, member 3
|
|
chr3_-_101077605
|
0.673
|
NM_014890
|
FILIP1L
|
filamin A interacting protein 1-like
|
|
chr3_+_112743615
|
0.672
|
NM_005816
NM_198196
|
CD96
|
CD96 molecule
|
|
chr7_-_116854765
|
0.671
|
NM_130768
|
ASZ1
|
ankyrin repeat, SAM and basic leucine zipper domain containing 1
|
|
chr13_+_51933228
|
0.670
|
|
CKAP2
|
cytoskeleton associated protein 2
|
|
chr4_+_71235259
|
0.668
|
NM_033122
|
CABS1
|
calcium-binding protein, spermatid-specific 1
|
|
chr2_-_215953818
|
0.667
|
|
FN1
|
fibronectin 1
|
|
chr1_-_90955365
|
0.665
|
NM_020063
|
BARHL2
|
BarH-like homeobox 2
|
|
chr17_+_59244363
|
0.662
|
|
DDX42
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 42
|
|
chr17_-_36074812
|
0.658
|
NM_152349
|
KRT222
|
keratin 222
|
|
chr2_-_180319013
|
0.657
|
NM_001113397
|
ZNF385B
|
zinc finger protein 385B
|
|
chr3_-_121117647
|
0.656
|
|
GSK3B
|
glycogen synthase kinase 3 beta
|
|
chr4_-_64957595
|
0.655
|
NM_001010874
|
TECRL
|
trans-2,3-enoyl-CoA reductase-like
|
|
chr3_+_189425886
|
0.654
|
NM_001167671
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr1_+_74473672
|
0.653
|
NM_015978
|
TNNI3K
|
TNNI3 interacting kinase
|
|
chr6_-_127882192
|
0.651
|
NM_001012279
|
C6orf174
|
chromosome 6 open reading frame 174
|
|
chr12_-_98072602
|
0.646
|
NM_020140
NM_181670
|
ANKS1B
|
ankyrin repeat and sterile alpha motif domain containing 1B
|
|
chr9_+_106371269
|
0.645
|
NM_001004483
|
OR13C8
|
olfactory receptor, family 13, subfamily C, member 8
|
|
chr1_-_202730135
|
0.638
|
|
PIK3C2B
|
phosphoinositide-3-kinase, class 2, beta polypeptide
|
|
chr10_+_115514567
|
0.636
|
NM_182601
|
C10orf81
|
chromosome 10 open reading frame 81
|
|
chr7_-_122126422
|
0.635
|
NM_139175
|
RNF133
|
ring finger protein 133
|
|
chr3_+_112743729
|
0.632
|
|
CD96
|
CD96 molecule
|
|
chr9_-_40782062
|
0.630
|
NM_033160
|
ZNF658
|
zinc finger protein 658
|