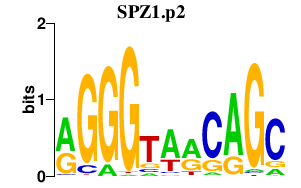
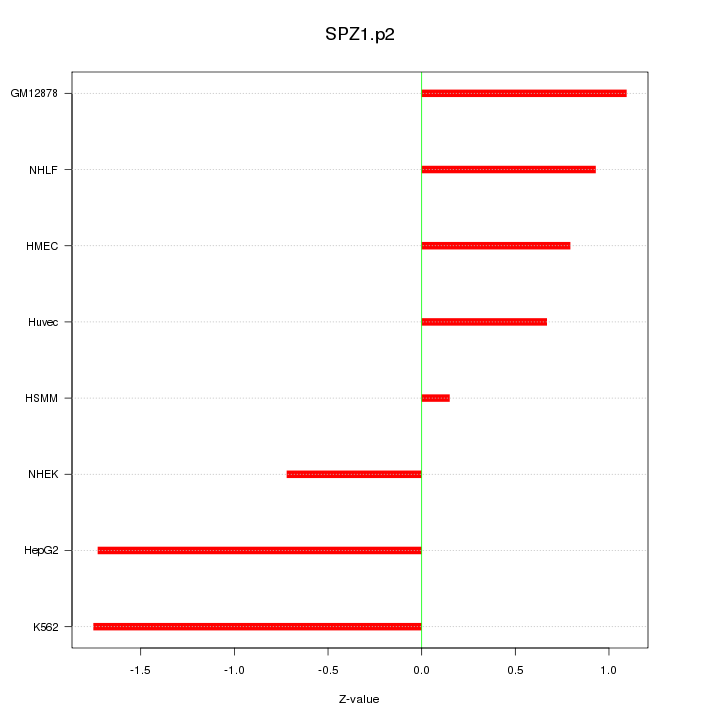
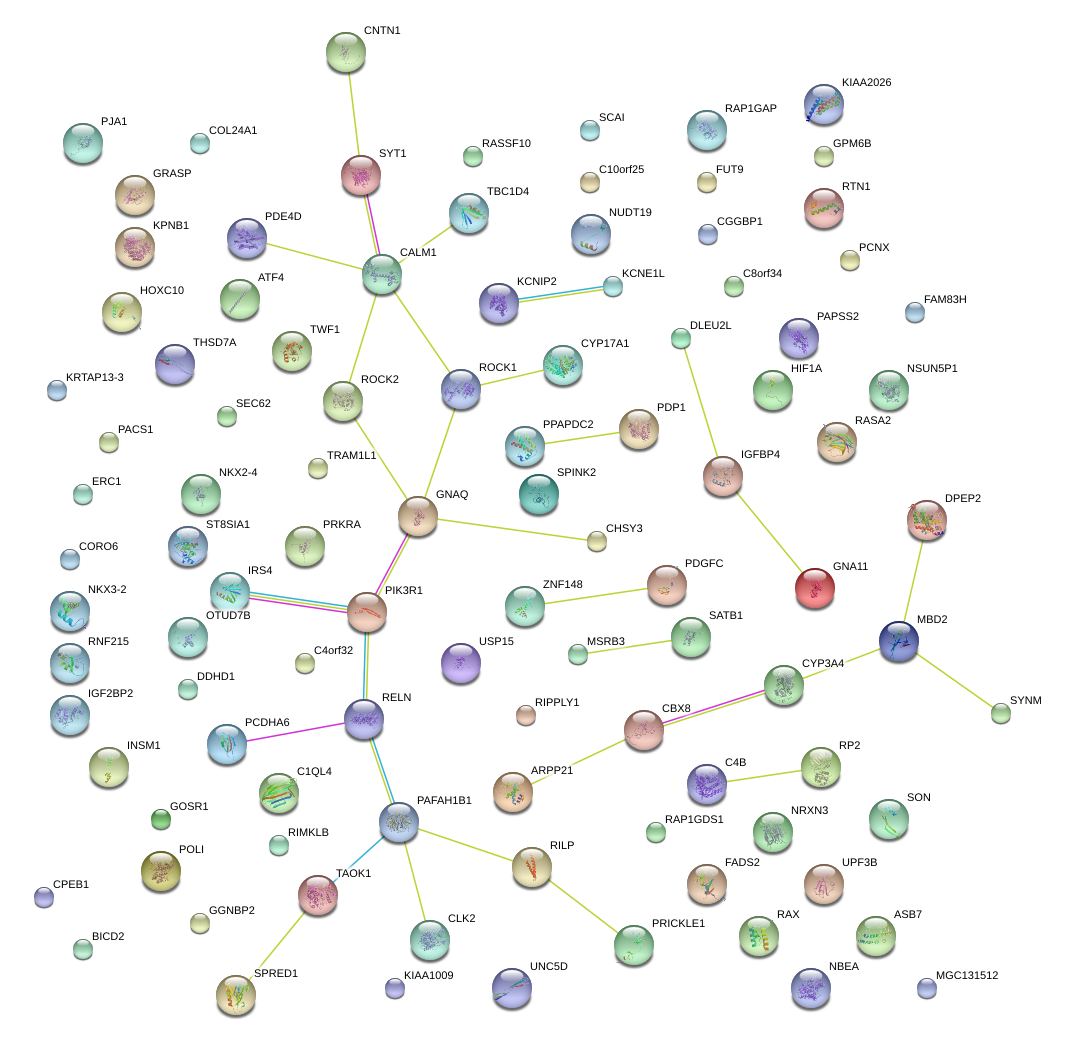
|
chr3_-_88190737
|
1.411
|
NM_001008390
NM_003663
|
CGGBP1
|
CGG triplet repeat binding protein 1
|
|
chr9_-_5997891
|
0.928
|
NM_001017969
|
KIAA2026
|
KIAA2026
|
|
chr11_+_12987455
|
0.782
|
NM_001080521
|
RASSF10
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 10
|
|
chr12_+_50686990
|
0.782
|
NM_181711
|
GRASP
|
GRP1 (general receptor for phosphoinositides 1)-associated scaffold protein
|
|
chr8_+_94998258
|
0.758
|
NM_018444
NM_001161780
NM_001161779
NM_001161781
|
PDP1
|
pyruvate dehyrogenase phosphatase catalytic subunit 1
|
|
chr17_+_31975611
|
0.735
|
|
GGNBP2
|
gametogenetin binding protein 2
|
|
chr12_+_52665196
|
0.713
|
NM_017409
|
HOXC10
|
homeobox C10
|
|
chr13_-_74953873
|
0.711
|
NM_014832
|
TBC1D4
|
TBC1 domain family, member 4
|
|
chr21_+_33844793
|
0.705
|
|
SON
|
SON DNA binding protein
|
|
chr1_-_86394695
|
0.689
|
NM_152890
|
COL24A1
|
collagen, type XXIV, alpha 1
|
|
chr10_-_104587069
|
0.639
|
NM_000102
|
CYP17A1
|
cytochrome P450, family 17, subfamily A, polypeptide 1
|
|
chr11_+_65594361
|
0.633
|
NM_018026
|
PACS1
|
phosphofurin acidic cluster sorting protein 1
|
|
chr12_+_63958949
|
0.622
|
NM_001193461
|
MSRB3
|
methionine sulfoxide reductase B3
|
|
chr13_+_34414390
|
0.621
|
NM_015678
|
NBEA
|
neurobeachin
|
|
chr17_+_35853177
|
0.571
|
NM_001552
|
IGFBP4
|
insulin-like growth factor binding protein 4
|
|
chr20_-_21326046
|
0.570
|
NM_033176
|
NKX2-4
|
NK2 homeobox 4
|
|
chr17_-_24973940
|
0.565
|
|
CORO6
|
coronin 6
|
|
chr17_+_2443983
|
0.561
|
|
PAFAH1B1
|
platelet-activating factor acetylhydrolase 1b, regulatory subunit 1 (45kDa)
|
|
chr20_+_20296744
|
0.555
|
NM_002196
|
INSM1
|
insulinoma-associated 1
|
|
chr3_-_18441763
|
0.541
|
NM_001195470
NM_002971
|
SATB1
|
SATB homeobox 1
|
|
chr10_-_103593529
|
0.534
|
NM_014591
NM_173191
NM_173192
NM_173193
NM_173195
NM_173197
|
KCNIP2
|
Kv channel interacting protein 2
|
|
chr7_-_103417198
|
0.530
|
NM_005045
NM_173054
|
RELN
|
reelin
|
|
chr1_-_148249152
|
0.529
|
NM_020205
|
OTUD7B
|
OTU domain containing 7B
|
|
chr3_+_171167243
|
0.513
|
NM_003262
|
SEC62
|
SEC62 homolog (S. cerevisiae)
|
|
chr12_-_22378833
|
0.508
|
NM_003034
|
ST8SIA1
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 1
|
|
chr10_+_89409332
|
0.506
|
NM_001015880
NM_004670
|
PAPSS2
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2
|
|
chr22_-_29113301
|
0.505
|
NM_001017981
|
RNF215
|
ring finger protein 215
|
|
chrX_-_118870936
|
0.505
|
NM_023010
NM_080632
|
UPF3B
|
UPF3 regulator of nonsense transcripts homolog B (yeast)
|
|
chr1_-_21868380
|
0.495
|
NM_001145657
NM_002885
|
RAP1GAP
|
RAP1 GTPase activating protein
|
|
chr3_-_62835601
|
0.489
|
|
|
|
|
chr12_+_970613
|
0.487
|
NM_178039
NM_178040
|
ERC1
|
ELKS/RAB6-interacting/CAST family member 1
|
|
chr13_-_49597776
|
0.459
|
|
DLEU2
|
deleted in lymphocytic leukemia 2 (non-protein coding)
|
|
chr12_+_39372456
|
0.458
|
NM_001843
NM_175038
|
CNTN1
|
contactin 1
|
|
chr4_-_13155135
|
0.455
|
NM_001189
|
NKX3-2
|
NK3 homeobox 2
|
|
chr7_-_99219639
|
0.451
|
NM_017460
|
CYP3A4
|
cytochrome P450, family 3, subfamily A, polypeptide 4
|
|
chr5_+_67547357
|
0.448
|
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha)
|
|
chr14_+_89934358
|
0.432
|
NM_001166106
|
CALM1
|
calmodulin 1 (phosphorylase kinase, delta)
|
|
chr6_+_96570565
|
0.430
|
NM_006581
|
FUT9
|
fucosyltransferase 9 (alpha (1,3) fucosyltransferase)
|
|
chr5_-_59225377
|
0.428
|
NM_001104631
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chrX_+_46581418
|
0.426
|
|
RP2
|
retinitis pigmentosa 2 (X-linked recessive)
|
|
chr14_-_59407103
|
0.425
|
NM_021136
|
RTN1
|
reticulon 1
|
|
chr18_-_55091596
|
0.424
|
NM_013435
|
RAX
|
retina and anterior neural fold homeobox
|
|
chr4_-_57382584
|
0.421
|
NM_021114
|
SPINK2
|
serine peptidase inhibitor, Kazal type 2 (acrosin-trypsin inhibitor)
|
|
chr15_+_97462675
|
0.420
|
NM_015286
NM_145728
|
SYNM
|
synemin, intermediate filament protein
|
|
chr5_+_129268352
|
0.417
|
NM_175856
|
CHSY3
|
chondroitin sulfate synthase 3
|
|
chr14_+_85069217
|
0.414
|
|
|
|
|
chr18_+_50049846
|
0.414
|
NM_007195
|
POLI
|
polymerase (DNA directed) iota
|
|
chrX_-_106033202
|
0.413
|
NM_001171706
NM_138382
|
RIPPLY1
|
ripply1 homolog (zebrafish)
|
|
chr4_+_99401543
|
0.405
|
NM_001100426
NM_001100427
NM_001100428
NM_001100429
NM_001100430
NM_021159
|
RAP1GDS1
|
RAP1, GTP-GDP dissociation stimulator 1
|
|
chr18_-_50004458
|
0.401
|
|
MBD2
|
methyl-CpG binding domain protein 2
|
|
chr11_+_61352348
|
0.398
|
|
FADS2
|
fatty acid desaturase 2
|
|
chr10_-_44816265
|
0.396
|
NM_001039380
|
C10orf25
|
chromosome 10 open reading frame 25
|
|
chr15_+_36332258
|
0.395
|
NM_152594
|
SPRED1
|
sprouty-related, EVH1 domain containing 1
|
|
chr9_-_126945536
|
0.393
|
NM_001144877
NM_173690
|
SCAI
|
suppressor of cancer cell invasion
|
|
chr14_+_78815406
|
0.387
|
NM_001105250
NM_138970
|
NRXN3
|
neurexin 3
|
|
chrX_-_13866451
|
0.385
|
|
GPM6B
|
glycoprotein M6B
|
|
chr12_+_8741746
|
0.383
|
NM_020734
|
RIMKLB
|
ribosomal modification protein rimK-like family member B
|
|
chr17_+_24742068
|
0.382
|
NM_020791
|
TAOK1
|
TAO kinase 1
|
|
chr7_-_11838260
|
0.380
|
NM_015204
|
THSD7A
|
thrombospondin, type I, domain containing 7A
|
|
chr6_-_84994032
|
0.378
|
NM_014895
|
KIAA1009
|
KIAA1009
|
|
chr14_-_52689680
|
0.378
|
|
DDHD1
|
DDHD domain containing 1
|
|
chr12_+_77782719
|
0.377
|
|
SYT1
|
synaptotagmin I
|
|
chrX_-_13866565
|
0.376
|
NM_001001994
|
GPM6B
|
glycoprotein M6B
|
|
chr2_-_11402150
|
0.375
|
NM_004850
|
ROCK2
|
Rho-associated, coiled-coil containing protein kinase 2
|
|
chr22_+_38246509
|
0.375
|
NM_001675
NM_182810
|
ATF4
|
activating transcription factor 4 (tax-responsive enhancer element B67)
|
|
chr8_+_35212505
|
0.374
|
NM_080872
|
UNC5D
|
unc-5 homolog D (C. elegans)
|
|
chr9_-_79835970
|
0.373
|
NM_002072
|
GNAQ
|
guanine nucleotide binding protein (G protein), q polypeptide
|
|
chr8_-_144887876
|
0.372
|
NM_198488
|
FAM83H
|
family with sequence similarity 83, member H
|
|
chr5_+_140286485
|
0.372
|
NM_018898
NM_031882
|
PCDHAC1
|
protocadherin alpha subfamily C, 1
|
|
chr14_-_52689570
|
0.369
|
|
DDHD1
|
DDHD domain containing 1
|
|
chrX_-_107866262
|
0.363
|
NM_003604
|
IRS4
|
insulin receptor substrate 4
|
|
chr17_-_1195096
|
0.359
|
|
PAFAH1B1
|
platelet-activating factor acetylhydrolase 1b, regulatory subunit 1 (45kDa)
|
|
chr12_-_42486314
|
0.357
|
NM_002822
|
TWF1
|
twinfilin, actin-binding protein, homolog 1 (Drosophila)
|
|
chr18_-_50004770
|
0.355
|
|
MBD2
|
methyl-CpG binding domain protein 2
|
|
chr17_-_75385371
|
0.355
|
NM_020649
|
CBX8
|
chromobox homolog 8
|
|
chr17_-_1499849
|
0.355
|
NM_031430
|
RILP
|
Rab interacting lysosomal protein
|
|
chrX_-_68301974
|
0.354
|
NM_001032396
NM_022368
NM_145119
|
PJA1
|
praja ring finger 1
|
|
chr8_+_68418622
|
0.352
|
|
|
|
|
chr17_+_43082273
|
0.352
|
NM_002265
|
KPNB1
|
karyopherin (importin) beta 1
|
|
chr8_+_69405510
|
0.346
|
NM_001195639
NM_052958
|
C8orf34
|
chromosome 8 open reading frame 34
|
|
chr15_-_81113676
|
0.346
|
NM_030594
|
CPEB1
|
cytoplasmic polyadenylation element binding protein 1
|
|
chr12_+_60940404
|
0.343
|
NM_006313
|
USP15
|
ubiquitin specific peptidase 15
|
|
chr15_+_98960277
|
0.342
|
NM_024708
NM_198243
|
ASB7
|
ankyrin repeat and SOCS box containing 7
|
|
chrX_-_108754906
|
0.339
|
NM_012282
|
KCNE1L
|
KCNE1-like
|
|
chr4_+_113286001
|
0.337
|
NM_152400
|
C4orf32
|
chromosome 4 open reading frame 32
|
|
chr9_-_94566823
|
0.336
|
NM_001003800
NM_015250
|
BICD2
|
bicaudal D homolog 2 (Drosophila)
|
|
chr17_+_25828546
|
0.335
|
NM_001007024
NM_001007025
NM_004871
|
GOSR1
|
golgi SNAP receptor complex member 1
|
|
chr14_+_61231795
|
0.335
|
NM_001530
NM_181054
|
HIF1A
|
hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor)
|
|
chr3_+_214668
|
0.333
|
|
|
|
|
chr4_-_118226119
|
0.330
|
NM_152402
|
TRAM1L1
|
translocation associated membrane protein 1-like 1
|
|
chr21_-_30720100
|
0.329
|
NM_181622
|
KRTAP13-3
|
keratin associated protein 13-3
|
|
chr3_-_187025438
|
0.329
|
|
IGF2BP2
|
insulin-like growth factor 2 mRNA binding protein 2
|
|
chrX_-_13866366
|
0.328
|
|
|
|
|
chr3_-_126576693
|
0.328
|
NM_021964
|
ZNF148
|
zinc finger protein 148
|
|
chr12_-_48017223
|
0.323
|
NM_001008223
|
C1QL4
|
complement component 1, q subcomponent-like 4
|
|
chr3_+_142688493
|
0.320
|
NM_006506
|
RASA2
|
RAS p21 protein activator 2
|
|
chr4_-_158111995
|
0.320
|
NM_016205
|
PDGFC
|
platelet derived growth factor C
|
|
chr14_+_70444191
|
0.319
|
|
PCNX
|
pecanex homolog (Drosophila)
|
|
chr3_+_35656012
|
0.317
|
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa
|
|
chr5_-_41546384
|
0.316
|
NM_001005473
|
PLCXD3
|
phosphatidylinositol-specific phospholipase C, X domain containing 3
|
|
chrX_+_46581368
|
0.315
|
|
RP2
|
retinitis pigmentosa 2 (X-linked recessive)
|
|
chr3_+_125786195
|
0.310
|
NM_007064
|
KALRN
|
kalirin, RhoGEF kinase
|
|
chr9_+_70509925
|
0.310
|
NM_003558
|
PIP5K1B
|
phosphatidylinositol-4-phosphate 5-kinase, type I, beta
|
|
chr14_-_70345485
|
0.309
|
NM_033141
|
MAP3K9
|
mitogen-activated protein kinase kinase kinase 9
|
|
chr3_-_151904408
|
0.306
|
NM_152394
|
FAM194A
|
family with sequence similarity 194, member A
|
|
chr11_+_12088639
|
0.305
|
NM_014632
|
MICAL2
|
microtubule associated monoxygenase, calponin and LIM domain containing 2
|
|
chr3_-_107070400
|
0.302
|
NM_170662
|
CBLB
|
Cas-Br-M (murine) ecotropic retroviral transforming sequence b
|
|
chr8_-_25371663
|
0.301
|
|
KCTD9
|
potassium channel tetramerisation domain containing 9
|
|
chr10_+_105243724
|
0.301
|
NM_004210
|
NEURL
|
neuralized homolog (Drosophila)
|
|
chr8_-_125454097
|
0.301
|
NM_194291
|
TMEM65
|
transmembrane protein 65
|
|
chr3_+_112273352
|
0.299
|
|
PVRL3
|
poliovirus receptor-related 3
|
|
chr11_+_45900731
|
0.298
|
|
GYLTL1B
|
glycosyltransferase-like 1B
|
|
chr7_-_37923040
|
0.295
|
NM_003014
|
SFRP4
|
secreted frizzled-related protein 4
|
|
chr14_+_69993969
|
0.294
|
NM_003813
|
ADAM21
|
ADAM metallopeptidase domain 21
|
|
chr20_+_17498507
|
0.289
|
NM_001011546
NM_006870
|
DSTN
|
destrin (actin depolymerizing factor)
|
|
chr11_+_27019084
|
0.288
|
NM_003986
|
BBOX1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1
|
|
chr1_+_69806617
|
0.287
|
|
LRRC7
|
leucine rich repeat containing 7
|
|
chr13_-_45910325
|
0.286
|
|
C13orf18
|
chromosome 13 open reading frame 18
|
|
chr9_-_19778590
|
0.284
|
NM_001193288
NM_020344
|
SLC24A2
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 2
|
|
chrX_+_46581290
|
0.283
|
NM_006915
|
RP2
|
retinitis pigmentosa 2 (X-linked recessive)
|
|
chr2_-_51112743
|
0.283
|
|
NRXN1
|
neurexin 1
|
|
chrX_-_132947290
|
0.281
|
NM_001164617
NM_001164618
NM_001164619
NM_004484
|
GPC3
|
glypican 3
|
|
chr8_+_79740813
|
0.280
|
NM_016010
|
FAM164A
|
family with sequence similarity 164, member A
|
|
chr19_-_54350457
|
0.280
|
NM_002152
|
HRC
|
histidine rich calcium binding protein
|
|
chr12_+_7233536
|
0.279
|
NM_001131023
NM_001131024
NM_001131025
|
PEX5
|
peroxisomal biogenesis factor 5
|
|
chr12_-_6852709
|
0.278
|
NM_001146316
NM_032641
|
SPSB2
|
splA/ryanodine receptor domain and SOCS box containing 2
|
|
chr9_+_70107602
|
0.278
|
NM_199135
|
FOXD4L3
FOXD4L6
|
forkhead box D4-like 3
forkhead box D4-like 6
|
|
chr5_+_139473873
|
0.277
|
NM_005859
|
PURA
|
purine-rich element binding protein A
|
|
chr15_+_92575767
|
0.277
|
|
MCTP2
|
multiple C2 domains, transmembrane 2
|
|
chr14_+_89597860
|
0.276
|
NM_022054
|
KCNK13
|
potassium channel, subfamily K, member 13
|
|
chr11_+_33236435
|
0.275
|
|
HIPK3
|
homeodomain interacting protein kinase 3
|
|
chr3_+_162697289
|
0.275
|
NM_001080440
|
OTOL1
|
otolin 1 homolog (zebrafish)
|
|
chr5_+_113726203
|
0.274
|
|
KCNN2
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 2
|
|
chr12_-_129889569
|
0.274
|
NM_001980
NM_194356
|
STX2
|
syntaxin 2
|
|
chr5_-_111782637
|
0.273
|
NM_022140
|
EPB41L4A
|
erythrocyte membrane protein band 4.1 like 4A
|
|
chrX_-_132947141
|
0.272
|
|
GPC3
|
glypican 3
|
|
chr10_-_98263434
|
0.272
|
NM_012465
|
TLL2
|
tolloid-like 2
|
|
chr6_-_80713589
|
0.272
|
NM_022726
|
ELOVL4
|
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 4
|
|
chr12_-_55134692
|
0.272
|
NM_012064
|
MIP
|
major intrinsic protein of lens fiber
|
|
chr17_-_7036490
|
0.271
|
|
|
|
|
chr9_+_115678365
|
0.266
|
NM_133374
|
ZNF618
|
zinc finger protein 618
|
|
chr12_+_26003218
|
0.266
|
NM_001164748
NM_007211
|
RASSF8
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8
|
|
chr6_-_110607496
|
0.266
|
|
WASF1
|
WAS protein family, member 1
|
|
chr9_-_35105844
|
0.266
|
NM_025182
|
KIAA1539
|
KIAA1539
|
|
chr10_+_76541351
|
0.265
|
NM_001174156
NM_144660
|
SAMD8
|
sterile alpha motif domain containing 8
|
|
chr10_+_121475583
|
0.265
|
NM_014937
|
INPP5F
|
inositol polyphosphate-5-phosphatase F
|
|
chr14_-_64416307
|
0.264
|
|
SPTB
|
spectrin, beta, erythrocytic
|
|
chr15_-_81412354
|
0.264
|
NM_004839
NM_199330
NM_199331
NM_199332
|
HOMER2
|
homer homolog 2 (Drosophila)
|
|
chr1_+_26896519
|
0.263
|
|
ARID1A
|
AT rich interactive domain 1A (SWI-like)
|
|
chr13_+_41520888
|
0.263
|
NM_152910
NM_178009
|
DGKH
|
diacylglycerol kinase, eta
|
|
chr5_-_93102952
|
0.262
|
NM_153216
|
POU5F2
|
POU domain class 5, transcription factor 2
|
|
chr9_-_85342868
|
0.261
|
NM_174938
|
FRMD3
|
FERM domain containing 3
|
|
chr12_-_52980914
|
0.258
|
|
NFE2
|
nuclear factor (erythroid-derived 2), 45kDa
|
|
chr5_-_88016397
|
0.257
|
|
LOC645323
|
hypothetical LOC645323
|
|
chr18_-_38949481
|
0.256
|
NM_002930
|
RIT2
|
Ras-like without CAAX 2
|
|
chr1_+_166414839
|
0.256
|
|
TIPRL
|
TIP41, TOR signaling pathway regulator-like (S. cerevisiae)
|
|
chrX_+_23262905
|
0.255
|
NM_173495
|
PTCHD1
|
patched domain containing 1
|
|
chr1_-_11663762
|
0.255
|
NM_006341
|
MAD2L2
|
MAD2 mitotic arrest deficient-like 2 (yeast)
|
|
chr3_-_48204804
|
0.255
|
NM_001789
NM_201567
|
CDC25A
|
cell division cycle 25 homolog A (S. pombe)
|
|
chr9_-_34512972
|
0.255
|
NM_198573
|
ENHO
|
energy homeostasis associated
|
|
chr17_-_2153763
|
0.255
|
NM_017575
|
SMG6
|
Smg-6 homolog, nonsense mediated mRNA decay factor (C. elegans)
|
|
chr5_-_114543632
|
0.254
|
NM_001017397
NM_001017398
NM_018700
|
TRIM36
|
tripartite motif containing 36
|
|
chr10_+_98582648
|
0.254
|
NM_032440
|
LCOR
|
ligand dependent nuclear receptor corepressor
|
|
chr5_-_158569067
|
0.254
|
|
RNF145
|
ring finger protein 145
|
|
chr6_-_70563489
|
0.253
|
|
LMBRD1
|
LMBR1 domain containing 1
|
|
chr12_+_48303607
|
0.252
|
NM_012272
|
PRPF40B
|
PRP40 pre-mRNA processing factor 40 homolog B (S. cerevisiae)
|
|
chr2_-_220144254
|
0.252
|
|
OBSL1
|
obscurin-like 1
|
|
chr12_-_78608154
|
0.252
|
|
PAWR
|
PRKC, apoptosis, WT1, regulator
|
|
chrX_-_83329562
|
0.252
|
NM_014496
|
RPS6KA6
|
ribosomal protein S6 kinase, 90kDa, polypeptide 6
|
|
chr4_-_44145266
|
0.250
|
|
KCTD8
|
potassium channel tetramerisation domain containing 8
|
|
chr14_+_64949447
|
0.249
|
|
FUT8
|
fucosyltransferase 8 (alpha (1,6) fucosyltransferase)
|
|
chr8_+_22513043
|
0.249
|
NM_001013842
NM_001198827
NM_173686
|
C8orf58
|
chromosome 8 open reading frame 58
|
|
chr3_-_143350898
|
0.249
|
NM_001178138
NM_001178139
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2)
|
|
chr15_-_72952648
|
0.248
|
NM_005697
|
SCAMP2
|
secretory carrier membrane protein 2
|
|
chr17_+_43082557
|
0.247
|
|
KPNB1
|
karyopherin (importin) beta 1
|
|
chr3_-_187025501
|
0.247
|
NM_001007225
NM_006548
|
IGF2BP2
|
insulin-like growth factor 2 mRNA binding protein 2
|
|
chr1_+_69806543
|
0.246
|
|
|
|
|
chr9_-_121171400
|
0.246
|
NM_014618
|
DBC1
|
deleted in bladder cancer 1
|
|
chr12_+_6907646
|
0.245
|
NM_001940
|
ATN1
|
atrophin 1
|
|
chr4_+_142777172
|
0.245
|
NM_000585
NM_172174
|
IL15
|
interleukin 15
|
|
chr15_+_83724743
|
0.245
|
NM_006738
NM_007200
|
AKAP13
|
A kinase (PRKA) anchor protein 13
|
|
chr18_-_22383066
|
0.244
|
NM_001136205
|
KCTD1
|
potassium channel tetramerisation domain containing 1
|
|
chr1_-_49014993
|
0.243
|
NM_024603
|
BEND5
|
BEN domain containing 5
|
|
chr5_+_17270781
|
0.242
|
|
BASP1
|
brain abundant, membrane attached signal protein 1
|
|
chr13_+_30089829
|
0.241
|
NM_005800
|
USPL1
|
ubiquitin specific peptidase like 1
|
|
chr6_+_124166767
|
0.240
|
NM_001040214
NM_153355
|
NKAIN2
|
Na+/K+ transporting ATPase interacting 2
|
|
chr11_-_94603857
|
0.240
|
NM_144665
|
SESN3
|
sestrin 3
|
|
chr1_-_32479785
|
0.239
|
|
MTMR9LP
|
myotubularin related protein 9-like, pseudogene
|
|
chr10_-_97040661
|
0.239
|
NM_020992
|
PDLIM1
|
PDZ and LIM domain 1
|
|
chr3_-_188870799
|
0.239
|
NM_001048
|
SST
|
somatostatin
|
|
chr14_+_70178126
|
0.237
|
NM_015351
|
TTC9
|
tetratricopeptide repeat domain 9
|
|
chr8_+_76482661
|
0.237
|
|
HNF4G
|
hepatocyte nuclear factor 4, gamma
|
|
chr11_-_10272256
|
0.236
|
NM_030962
|
SBF2
|
SET binding factor 2
|
|
chr6_+_72653420
|
0.236
|
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr10_-_104943000
|
0.236
|
NM_001134373
NM_012229
|
NT5C2
|
5'-nucleotidase, cytosolic II
|
|
chr6_-_70563564
|
0.236
|
NM_018368
|
LMBRD1
|
LMBR1 domain containing 1
|
|
chr5_+_96023926
|
0.236
|
|
CAST
|
calpastatin
|
|
chr3_+_113180517
|
0.235
|
NM_018394
|
ABHD10
|
abhydrolase domain containing 10
|
|
chr6_-_30151526
|
0.235
|
NM_170769
NM_025236
|
RNF39
|
ring finger protein 39
|
|
chr14_+_60858151
|
0.235
|
NM_006255
|
PRKCH
|
protein kinase C, eta
|