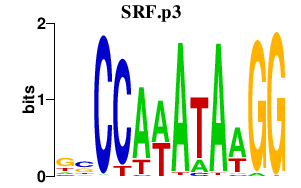
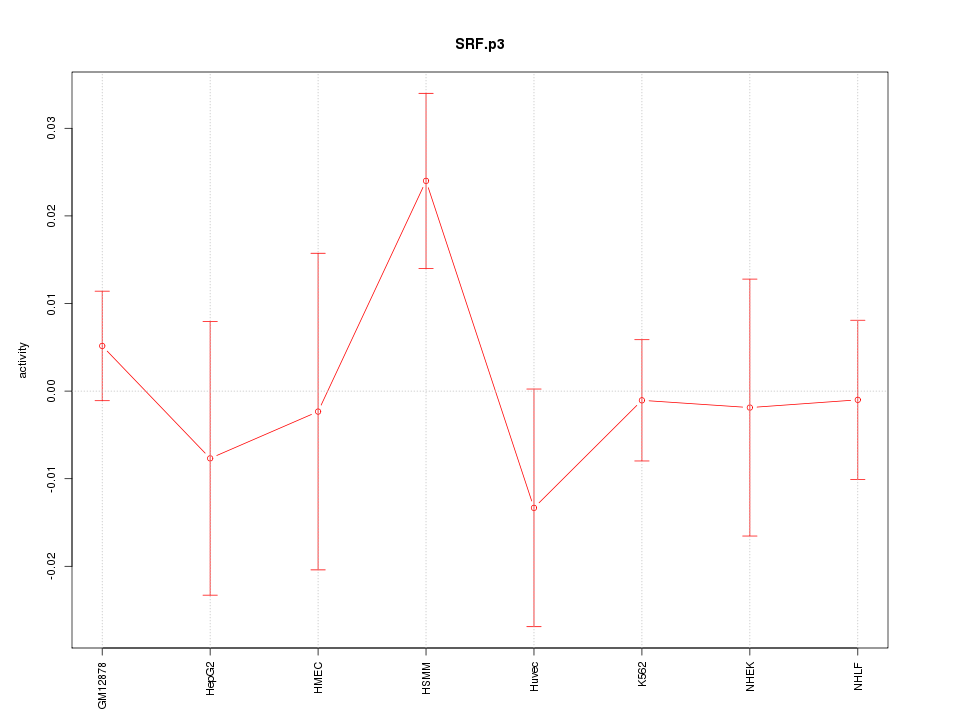
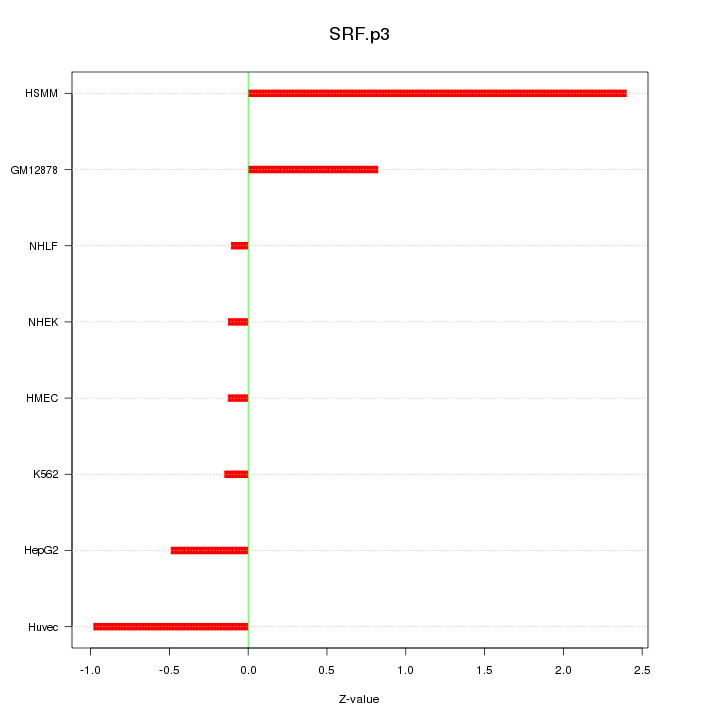
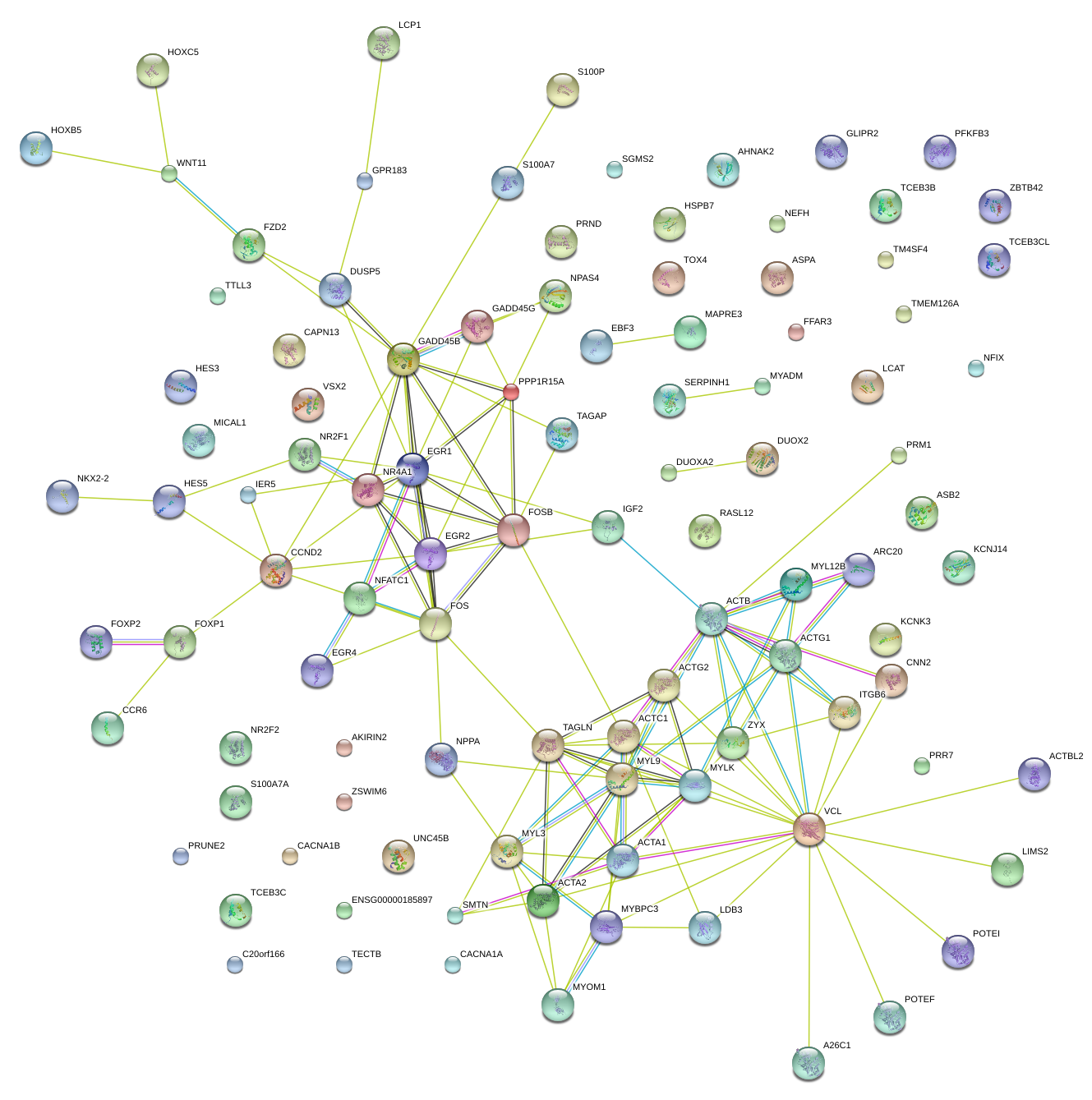
|
chr5_+_137829065
|
17.380
|
|
EGR1
|
early growth response 1
|
|
chr5_+_137829077
|
15.395
|
NM_001964
|
EGR1
|
early growth response 1
|
|
chr17_+_30498948
|
4.316
|
NM_001033576
NM_173167
|
UNC45B
|
unc-45 homolog B (C. elegans)
|
|
chr10_-_64246129
|
3.720
|
NM_000399
NM_001136177
NM_001136179
|
EGR2
|
early growth response 2
|
|
chr20_+_34603290
|
3.599
|
NM_006097
NM_181526
|
MYL9
|
myosin, light chain 9, regulatory
|
|
chr12_+_50731457
|
3.181
|
NM_002135
NM_173157
|
NR4A1
|
nuclear receptor subfamily 4, group A, member 1
|
|
chr20_+_34603311
|
3.057
|
|
MYL9
|
myosin, light chain 9, regulatory
|
|
chr12_+_52713098
|
2.852
|
NM_018953
|
HOXC5
|
homeobox C5
|
|
chr10_-_64246080
|
2.460
|
|
EGR2
|
early growth response 2
|
|
chr19_+_2427122
|
2.435
|
NM_015675
|
GADD45B
|
growth arrest and DNA-damage-inducible, beta
|
|
chr10_+_112247614
|
2.427
|
NM_004419
|
DUSP5
|
dual specificity phosphatase 5
|
|
chr9_+_91409746
|
2.175
|
NM_006705
|
GADD45G
|
growth arrest and DNA-damage-inducible, gamma
|
|
chr1_+_6226838
|
2.100
|
NM_001024598
|
HES3
|
hairy and enhancer of split 3 (Drosophila)
|
|
chr3_-_46879883
|
2.073
|
NM_000258
|
MYL3
|
myosin, light chain 3, alkali; ventricular, skeletal, slow
|
|
chr22_+_29810981
|
1.973
|
|
SMTN
|
smoothelin
|
|
chr2_+_73973600
|
1.964
|
NM_001615
|
ACTG2
|
actin, gamma 2, smooth muscle, enteric
|
|
chr2_-_30883775
|
1.889
|
NM_144575
|
CAPN13
|
calpain 13
|
|
chr20_-_21442663
|
1.788
|
NM_002509
|
NKX2-2
|
NK2 homeobox 2
|
|
chr2_+_73973640
|
1.764
|
|
ACTG2
|
actin, gamma 2, smooth muscle, enteric
|
|
chr19_+_12995782
|
1.723
|
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor)
|
|
chr11_+_116575249
|
1.619
|
NM_001001522
NM_003186
|
TAGLN
|
transgelin
|
|
chr19_+_50663089
|
1.594
|
NM_001114171
NM_006732
|
FOSB
|
FBJ murine osteosarcoma viral oncogene homolog B
|
|
chr9_+_36126710
|
1.412
|
NM_022343
|
GLIPR2
|
GLI pathogenesis-related 2
|
|
chr9_+_36126667
|
1.314
|
|
GLIPR2
|
GLI pathogenesis-related 2
|
|
chr19_+_53650577
|
1.302
|
NM_013348
|
KCNJ14
|
potassium inwardly-rectifying channel, subfamily J, member 14
|
|
chr14_+_73775927
|
1.279
|
NM_182894
|
VSX2
|
visual system homeobox 2
|
|
chr20_+_4650499
|
1.188
|
NM_012409
|
PRND
|
prion protein 2 (dublet)
|
|
chr9_+_139892049
|
1.096
|
NM_000718
|
CACNA1B
|
calcium channel, voltage-dependent, N type, alpha 1B subunit
|
|
chr11_+_74950893
|
1.086
|
|
SERPINH1
|
serpin peptidase inhibitor, clade H (heat shock protein 47), member 1, (collagen binding protein 1)
|
|
chr11_-_47330797
|
1.035
|
NM_000256
|
MYBPC3
|
myosin binding protein C, cardiac
|
|
chr16_-_11282595
|
0.969
|
NM_002761
|
PRM1
|
protamine 1
|
|
chr5_+_60663842
|
0.941
|
NM_020928
|
ZSWIM6
|
zinc finger, SWIM-type containing 6
|
|
chr15_+_94670160
|
0.938
|
NM_001145155
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr13_-_45640679
|
0.935
|
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin)
|
|
chr2_+_87536106
|
0.918
|
|
NCRNA00152
|
non-protein coding RNA 152
|
|
chr17_+_3326045
|
0.914
|
NM_000049
|
ASPA
|
aspartoacylase
|
|
chr14_-_93512818
|
0.905
|
|
ASB2
|
ankyrin repeat and SOCS box containing 2
|
|
chr2_-_160765008
|
0.892
|
|
ITGB6
|
integrin, beta 6
|
|
chr11_+_65945050
|
0.854
|
NM_178864
|
NPAS4
|
neuronal PAS domain protein 4
|
|
chr1_-_11830328
|
0.802
|
NM_006172
|
NPPA
|
natriuretic peptide A
|
|
chr2_-_128149112
|
0.800
|
NM_001136037
NM_001161403
|
LIMS2
|
LIM and senescent cell antigen-like domains 2
|
|
chr6_-_159386007
|
0.796
|
NM_054114
NM_138810
NM_152133
|
TAGAP
|
T-cell activation RhoGTPase activating protein
|
|
chr17_-_44026042
|
0.769
|
NM_002147
|
HOXB5
|
homeobox B5
|
|
chr14_+_104338423
|
0.753
|
|
ZBTB42
|
zinc finger and BTB domain containing 42
|
|
chr10_+_75427853
|
0.751
|
NM_003373
NM_014000
|
VCL
|
vinculin
|
|
chr3_-_124821868
|
0.744
|
|
MYLK
|
myosin light chain kinase
|
|
chr13_-_98757634
|
0.731
|
NM_004951
|
GPR183
|
G protein-coupled receptor 183
|
|
chr1_-_227636462
|
0.724
|
NM_001100
|
ACTA1
|
actin, alpha 1, skeletal muscle
|
|
chr15_+_43193810
|
0.716
|
NM_207581
|
DUOXA2
|
dual oxidase maturation factor 2
|
|
chr9_-_78710698
|
0.714
|
NM_015225
|
PRUNE2
|
prune homolog 2 (Drosophila)
|
|
chr17_-_44026321
|
0.705
|
|
HOXB5
|
homeobox B5
|
|
chr2_-_73373956
|
0.698
|
NM_001965
|
EGR4
|
early growth response 4
|
|
chr6_-_109882116
|
0.683
|
|
MICAL1
|
microtubule associated monoxygenase, calponin and LIM domain containing 1
|
|
chr15_-_63147350
|
0.674
|
NM_016563
|
RASL12
|
RAS-like, family 12
|
|
chr18_-_42804531
|
0.671
|
NM_145653
|
TCEB3C
|
transcription elongation factor B polypeptide 3C (elongin A3)
|
|
chr17_+_39990124
|
0.652
|
NM_001466
|
FZD2
|
frizzled homolog 2 (Drosophila)
|
|
chr19_+_977582
|
0.616
|
|
CNN2
|
calponin 2
|
|
chr10_-_131652364
|
0.613
|
|
EBF3
|
early B-cell factor 3
|
|
chr3_-_71376919
|
0.601
|
|
FOXP1
|
forkhead box P1
|
|
chr11_-_75599433
|
0.599
|
|
WNT11
|
wingless-type MMTV integration site family, member 11
|
|
chr3_+_150675117
|
0.597
|
NM_004617
|
TM4SF4
|
transmembrane 4 L six family member 4
|
|
chr16_-_66535457
|
0.595
|
NM_000229
|
LCAT
|
lecithin-cholesterol acyltransferase
|
|
chr18_-_42804291
|
0.593
|
NM_001100817
|
TCEB3C
TCEB3CL
|
transcription elongation factor B polypeptide 3C (elongin A3)
transcription elongation factor B polypeptide 3C-like
|
|
chr4_+_109033868
|
0.593
|
NM_001136257
NM_152621
|
SGMS2
|
sphingomyelin synthase 2
|
|
chr4_+_6746466
|
0.559
|
NM_005980
|
S100P
|
S100 calcium binding protein P
|
|
chr1_-_2451543
|
0.559
|
NM_001010926
|
HES5
|
hairy and enhancer of split 5 (Drosophila)
|
|
chr12_+_4253143
|
0.551
|
NM_001759
|
CCND2
|
cyclin D2
|
|
chr7_+_142788480
|
0.551
|
NM_001010972
NM_003461
|
ZYX
|
zyxin
|
|
chr1_+_179324458
|
0.549
|
|
IER5
|
immediate early response 5
|
|
chr22_+_28206180
|
0.543
|
NM_021076
|
NEFH
|
neurofilament, heavy polypeptide
|
|
chr14_-_93512647
|
0.531
|
|
ASB2
|
ankyrin repeat and SOCS box containing 2
|
|
chr18_+_75388522
|
0.530
|
|
NFATC1
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1
|
|
chr6_-_88468368
|
0.529
|
|
AKIRIN2
|
akirin 2
|
|
chr19_+_40541327
|
0.528
|
NM_005304
|
FFAR3
|
free fatty acid receptor 3
|
|
chr7_+_142788521
|
0.515
|
|
ZYX
|
zyxin
|
|
chr19_+_12996368
|
0.511
|
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor)
|
|
chr14_-_104507862
|
0.498
|
|
AHNAK2
|
AHNAK nucleoprotein 2
|
|
chr10_+_88418243
|
0.493
|
NM_001080116
NM_001080114
NM_001080115
NM_001171611
NM_007078
|
LDB3
|
LIM domain binding 3
|
|
chr15_-_43193637
|
0.491
|
NM_014080
|
DUOX2
|
dual oxidase 2
|
|
chr7_+_142788569
|
0.484
|
|
ZYX
|
zyxin
|
|
chr19_+_977608
|
0.479
|
|
CNN2
|
calponin 2
|
|
chr14_+_74815233
|
0.476
|
NM_005252
|
FOS
|
FBJ murine osteosarcoma viral oncogene homolog
|
|
chr20_+_60558104
|
0.474
|
NM_178463
|
C20orf166
|
chromosome 20 open reading frame 166
|
|
chr7_+_113842284
|
0.470
|
NM_001172766
NM_014491
NM_148898
NM_148900
|
FOXP2
|
forkhead box P2
|
|
chr19_+_59064795
|
0.468
|
|
MYADM
|
myeloid-associated differentiation marker
|
|
chr5_+_169749068
|
0.465
|
|
|
|
|
chr5_+_176807931
|
0.457
|
NM_001174102
|
PRR7
|
proline rich 7 (synaptic)
|
|
chr1_-_16217025
|
0.454
|
|
HSPB7
|
heat shock 27kDa protein family, member 7 (cardiovascular)
|
|
chr19_+_977555
|
0.449
|
|
CNN2
|
calponin 2
|
|
chr6_+_167445284
|
0.442
|
NM_004367
|
CCR6
|
chemokine (C-C motif) receptor 6
|
|
chr7_+_142788517
|
0.435
|
|
ZYX
|
zyxin
|
|
chr18_-_3209846
|
0.434
|
|
MYOM1
|
myomesin 1, 185kDa
|
|
chr17_-_77094426
|
0.432
|
|
ACTG1
|
actin, gamma 1
|
|
chr19_+_54068300
|
0.430
|
|
PPP1R15A
|
protein phosphatase 1, regulatory (inhibitor) subunit 15A
|
|
chr7_+_142788566
|
0.429
|
|
ZYX
|
zyxin
|
|
chr10_+_6226884
|
0.420
|
|
PFKFB3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3
|
|
chr10_+_114033403
|
0.420
|
NM_058222
|
TECTB
|
tectorin beta
|
|
chr11_-_2118743
|
0.416
|
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chr2_+_26768894
|
0.410
|
NM_002246
|
KCNK3
|
potassium channel, subfamily K, member 3
|
|
chr10_+_6226909
|
0.410
|
|
PFKFB3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3
|
|
chr1_+_151655623
|
0.402
|
NM_176823
|
S100A7A
|
S100 calcium binding protein A7A
|
|
chr3_+_9809776
|
0.402
|
NM_001198780
|
ARPC4
|
actin related protein 2/3 complex, subunit 4, 20kDa
|
|
chr19_+_977624
|
0.399
|
|
CNN2
|
calponin 2
|
|
chr19_+_977601
|
0.392
|
|
CNN2
|
calponin 2
|
|
chr1_-_31618405
|
0.391
|
NM_004102
|
FABP3
|
fatty acid binding protein 3, muscle and heart (mammary-derived growth inhibitor)
|
|
chr20_+_32610161
|
0.387
|
NM_032514
|
MAP1LC3A
|
microtubule-associated protein 1 light chain 3 alpha
|
|
chr22_+_36412277
|
0.379
|
NM_024313
|
NOL12
|
nucleolar protein 12
|
|
chr17_-_46296376
|
0.379
|
NM_005749
|
TOB1
|
transducer of ERBB2, 1
|
|
chrX_+_99726445
|
0.376
|
NM_022144
|
TNMD
|
tenomodulin
|
|
chr14_-_105163301
|
0.372
|
|
IGHG4
|
immunoglobulin heavy constant gamma 4 (G4m marker)
|
|
chr12_-_46405477
|
0.371
|
NM_001172439
NM_001172440
NM_006025
|
ENDOU
|
endonuclease, polyU-specific
|
|
chr15_-_43193614
|
0.368
|
|
DUOX2
|
dual oxidase 2
|
|
chr19_+_977627
|
0.364
|
|
CNN2
|
calponin 2
|
|
chr7_+_142788730
|
0.361
|
|
ZYX
|
zyxin
|
|
chr22_+_38246509
|
0.359
|
NM_001675
NM_182810
|
ATF4
|
activating transcription factor 4 (tax-responsive enhancer element B67)
|
|
chr17_+_29707583
|
0.356
|
NM_005408
|
CCL13
|
chemokine (C-C motif) ligand 13
|
|
chr10_-_29963847
|
0.352
|
|
|
|
|
chr19_-_50912429
|
0.351
|
|
|
|
|
chr19_+_59064471
|
0.351
|
NM_001020821
NM_001020818
|
MYADM
|
myeloid-associated differentiation marker
|
|
chr10_+_88418147
|
0.344
|
NM_001171610
|
LDB3
|
LIM domain binding 3
|
|
chr11_+_43920385
|
0.344
|
NM_001145033
|
C11orf96
|
chromosome 11 open reading frame 96
|
|
chr2_+_87536174
|
0.338
|
|
NCRNA00152
|
non-protein coding RNA 152
|
|
chr17_-_77094375
|
0.335
|
NM_001614
|
ACTG1
|
actin, gamma 1
|
|
chr19_+_16039170
|
0.330
|
NM_001145160
|
TPM4
|
tropomyosin 4
|
|
chr16_+_29984647
|
0.327
|
|
ALDOA
|
aldolase A, fructose-bisphosphate
|
|
chr15_-_53369261
|
0.321
|
NM_183235
|
RAB27A
|
RAB27A, member RAS oncogene family
|
|
chr15_-_63147228
|
0.318
|
|
RASL12
|
RAS-like, family 12
|
|
chr20_-_62151216
|
0.314
|
NM_018419
|
SOX18
|
SRY (sex determining region Y)-box 18
|
|
chr5_-_178983241
|
0.312
|
|
HNRNPH1
|
heterogeneous nuclear ribonucleoprotein H1 (H)
|
|
chr7_+_113842470
|
0.306
|
|
FOXP2
|
forkhead box P2
|
|
chr22_-_41672990
|
0.306
|
NM_007229
|
PACSIN2
|
protein kinase C and casein kinase substrate in neurons 2
|
|
chr19_+_50702588
|
0.301
|
|
VASP
|
vasodilator-stimulated phosphoprotein
|
|
chr10_-_29963906
|
0.296
|
NM_021738
|
SVIL
|
supervillin
|
|
chr21_-_46528594
|
0.295
|
|
MCM3AP
|
minichromosome maintenance complex component 3 associated protein
|
|
chr11_-_8982107
|
0.293
|
NM_020645
|
NRIP3
|
nuclear receptor interacting protein 3
|
|
chr7_+_142788579
|
0.292
|
|
ZYX
|
zyxin
|
|
chr3_-_124822113
|
0.290
|
NM_053031
NM_053032
|
MYLK
|
myosin light chain kinase
|
|
chr14_-_73296643
|
0.290
|
NM_194278
|
C14orf43
|
chromosome 14 open reading frame 43
|
|
chr3_-_9809357
|
0.288
|
NM_006354
NM_133480
|
TADA3
|
transcriptional adaptor 3
|
|
chr17_+_16259636
|
0.286
|
|
TRPV2
|
transient receptor potential cation channel, subfamily V, member 2
|
|
chr17_+_16259573
|
0.284
|
NM_016113
|
TRPV2
|
transient receptor potential cation channel, subfamily V, member 2
|
|
chr1_-_89303478
|
0.282
|
NM_002053
|
GBP1
|
guanylate binding protein 1, interferon-inducible, 67kDa
|
|
chr10_+_6226764
|
0.278
|
NM_001145443
|
PFKFB3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3
|
|
chr15_+_31390454
|
0.277
|
NM_001036
|
RYR3
|
ryanodine receptor 3
|
|
chr6_+_12824873
|
0.274
|
|
PHACTR1
|
phosphatase and actin regulator 1
|
|
chr10_-_44794146
|
0.271
|
NM_007021
|
C10orf10
|
chromosome 10 open reading frame 10
|
|
chr22_+_36334400
|
0.267
|
NM_001001560
NM_001001561
NM_001172687
NM_013365
|
GGA1
|
golgi-associated, gamma adaptin ear containing, ARF binding protein 1
|
|
chr1_-_925332
|
0.265
|
NM_001142467
NM_021170
|
HES4
|
hairy and enhancer of split 4 (Drosophila)
|
|
chr15_+_72006015
|
0.264
|
|
LOXL1
|
lysyl oxidase-like 1
|
|
chr17_+_38256710
|
0.263
|
NM_003734
|
AOC3
|
amine oxidase, copper containing 3 (vascular adhesion protein 1)
|
|
chr17_-_34603577
|
0.262
|
|
CACNB1
|
calcium channel, voltage-dependent, beta 1 subunit
|
|
chr15_-_57452361
|
0.262
|
NM_004998
|
MYO1E
|
myosin IE
|
|
chr9_-_133574928
|
0.255
|
NM_198679
|
RAPGEF1
|
Rap guanine nucleotide exchange factor (GEF) 1
|
|
chr15_-_57452357
|
0.254
|
|
MYO1E
|
myosin IE
|
|
chr8_+_24827173
|
0.254
|
NM_005382
|
NEFM
|
neurofilament, medium polypeptide
|
|
chr15_+_38430517
|
0.251
|
NM_001145643
|
PHGR1
|
proline/histidine/glycine-rich 1
|
|
chr16_+_29984611
|
0.251
|
|
ALDOA
|
aldolase A, fructose-bisphosphate
|
|
chr22_+_36334811
|
0.248
|
|
GGA1
|
golgi-associated, gamma adaptin ear containing, ARF binding protein 1
|
|
chr1_-_206151055
|
0.248
|
NM_001025109
NM_001773
|
CD34
|
CD34 molecule
|
|
chr3_-_187124290
|
0.247
|
|
TRA2B
|
transformer 2 beta homolog (Drosophila)
|
|
chr6_-_16869579
|
0.239
|
NM_000332
NM_001128164
|
ATXN1
|
ataxin 1
|
|
chr20_+_2224612
|
0.239
|
NM_003245
|
TGM3
|
transglutaminase 3 (E polypeptide, protein-glutamine-gamma-glutamyltransferase)
|
|
chr11_-_57038891
|
0.238
|
NM_001198810
|
SLC43A1
|
solute carrier family 43, member 1
|
|
chrX_-_11194015
|
0.237
|
NM_013423
|
ARHGAP6
|
Rho GTPase activating protein 6
|
|
chr9_+_115396020
|
0.237
|
NM_144489
|
RGS3
|
regulator of G-protein signaling 3
|
|
chr2_-_27384617
|
0.230
|
NM_003353
|
UCN
|
urocortin
|
|
chr22_+_36334622
|
0.225
|
|
GGA1
|
golgi-associated, gamma adaptin ear containing, ARF binding protein 1
|
|
chr6_+_44322764
|
0.222
|
NM_007355
|
HSP90AB1
|
heat shock protein 90kDa alpha (cytosolic), class B member 1
|
|
chr5_+_131424245
|
0.216
|
NM_000588
|
IL3
|
interleukin 3 (colony-stimulating factor, multiple)
|
|
chr18_-_63333487
|
0.215
|
|
DSEL
|
dermatan sulfate epimerase-like
|
|
chr19_-_54618410
|
0.214
|
NM_178449
|
PTH2
|
parathyroid hormone 2
|
|
chr17_-_38385511
|
0.213
|
NM_025267
|
AARSD1
|
alanyl-tRNA synthetase domain containing 1
|
|
chr11_+_66070949
|
0.212
|
NM_001104
|
ACTN3
|
actinin, alpha 3
|
|
chr17_-_36938111
|
0.211
|
NM_002276
|
KRT19
|
keratin 19
|
|
chr10_-_98109048
|
0.211
|
NM_001040102
NM_001040103
NM_033207
|
OPALIN
|
oligodendrocytic myelin paranodal and inner loop protein
|
|
chr5_-_178983281
|
0.211
|
NM_005520
|
HNRNPH1
|
heterogeneous nuclear ribonucleoprotein H1 (H)
|
|
chr5_+_6799100
|
0.211
|
NM_001171806
|
PAPD7
|
PAP associated domain containing 7
|
|
chr12_+_48784057
|
0.211
|
NM_005276
|
GPD1
|
glycerol-3-phosphate dehydrogenase 1 (soluble)
|
|
chr15_+_94674849
|
0.210
|
NM_021005
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr6_+_151602787
|
0.207
|
NM_005100
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chr17_-_71017509
|
0.203
|
NM_001142643
|
CASKIN2
|
CASK interacting protein 2
|
|
chr19_+_46774370
|
0.197
|
NM_001098506
NM_033543
|
CEACAM21
|
carcinoembryonic antigen-related cell adhesion molecule 21
|
|
chr16_+_29984621
|
0.196
|
|
ALDOA
|
aldolase A, fructose-bisphosphate
|
|
chr20_-_61728866
|
0.196
|
|
GMEB2
|
glucocorticoid modulatory element binding protein 2
|
|
chr3_+_150675169
|
0.193
|
|
TM4SF4
|
transmembrane 4 L six family member 4
|
|
chr5_-_131907071
|
0.193
|
NM_000879
|
IL5
|
interleukin 5 (colony-stimulating factor, eosinophil)
|
|
chr1_-_925215
|
0.192
|
|
HES4
|
hairy and enhancer of split 4 (Drosophila)
|
|
chr18_-_42810446
|
0.192
|
NM_145653
|
TCEB3C
|
transcription elongation factor B polypeptide 3C (elongin A3)
|
|
chr19_-_51912136
|
0.189
|
NM_001079880
NM_001079881
NM_016457
|
PRKD2
|
protein kinase D2
|
|
chr20_+_60558369
|
0.188
|
|
C20orf166
|
chromosome 20 open reading frame 166
|
|
chr17_+_29636799
|
0.187
|
NM_002986
|
CCL11
|
chemokine (C-C motif) ligand 11
|
|
chr16_+_54247400
|
0.185
|
|
SLC6A2
|
solute carrier family 6 (neurotransmitter transporter, noradrenalin), member 2
|
|
chr2_-_160764819
|
0.181
|
NM_000888
|
ITGB6
|
integrin, beta 6
|
|
chr1_-_206151002
|
0.175
|
|
CD34
|
CD34 molecule
|
|
chr1_-_151699728
|
0.171
|
NM_002963
|
S100A7
|
S100 calcium binding protein A7
|
|
chr19_+_54068373
|
0.168
|
|
PPP1R15A
|
protein phosphatase 1, regulatory (inhibitor) subunit 15A
|
|
chr10_+_123912930
|
0.162
|
|
TACC2
|
transforming, acidic coiled-coil containing protein 2
|
|
chr3_+_63928459
|
0.161
|
NM_001128149
|
ATXN7
|
ataxin 7
|
|
chr9_-_97309301
|
0.160
|
NM_001083607
|
PTCH1
|
patched 1
|
|
chr19_-_63550712
|
0.159
|
|
A1BG
|
alpha-1-B glycoprotein
|