|
chr19_+_13090101
|
4.627
|
NM_052876
|
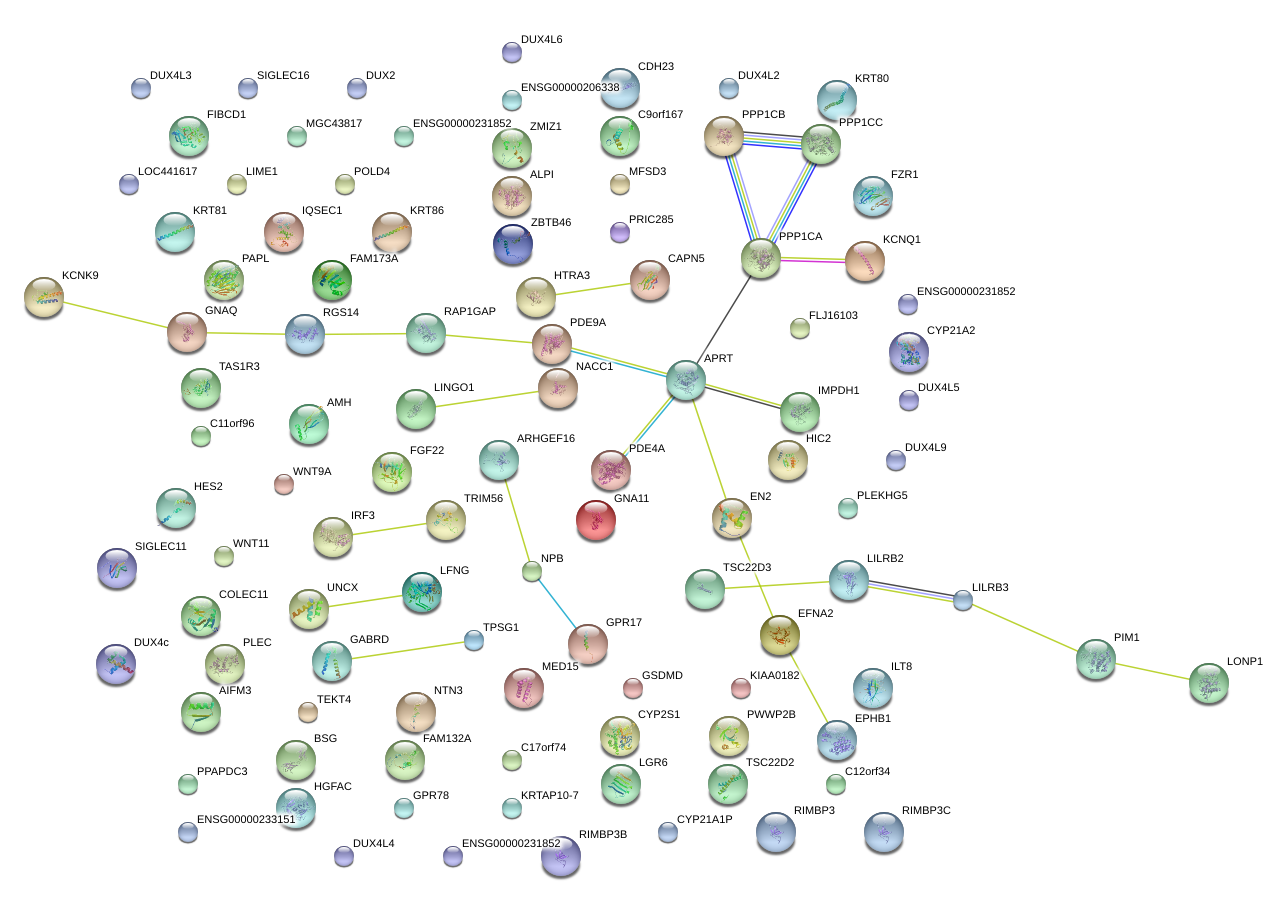
NACC1
|
nucleus accumbens associated 1, BEN and BTB (POZ) domain containing
|
|
chr16_-_87405817
|
3.249
|
NM_000485
NM_001030018
|
APRT
|
adenine phosphoribosyltransferase
|
|
chr22_+_20068039
|
3.056
|
NM_001128635
|
RIMBP3B
|
RIMS binding protein 3B
|
|
chr19_+_1236766
|
2.939
|
NM_001405
|
EFNA2
|
ephrin-A2
|
|
chr19_+_530466
|
2.847
|
|
BSG
|
basigin (Ok blood group)
|
|
chr20_-_61669550
|
2.739
|
NM_033405
|
PRIC285
|
peroxisomal proliferator-activated receptor A interacting complex 285
|
|
chr9_+_139292094
|
2.666
|
NM_017723
|
C9orf167
|
chromosome 9 open reading frame 167
|
|
chr6_+_32114060
|
2.566
|
NM_000500
NM_001128590
|
CYP21A2
|
cytochrome P450, family 21, subfamily A, polypeptide 2
|
|
chr3_-_12983959
|
2.428
|
NM_014869
|
IQSEC1
|
IQ motif and Sec7 domain 1
|
|
chr19_+_10392128
|
2.363
|
NM_001111307
|
PDE4A
|
phosphodiesterase 4A, cAMP-specific
|
|
chr22_+_19649417
|
2.206
|
NM_001018060
NM_144704
|
AIFM3
|
apoptosis-inducing factor, mitochondrion-associated, 3
|
|
chr7_+_1239078
|
2.197
|
NM_001080461
|
UNCX
|
UNC homeobox
|
|
chr19_+_590878
|
2.187
|
NM_020637
|
FGF22
|
fibroblast growth factor 22
|
|
chr12_-_50871953
|
2.158
|
NM_001081492
NM_182507
|
KRT80
|
keratin 80
|
|
chr8_+_144711619
|
2.154
|
NM_024736
|
GSDMD
|
gasdermin D
|
|
chr2_+_128119908
|
2.089
|
NM_001161415
NM_001161416
NM_001161417
NM_005291
|
GPR17
|
G protein-coupled receptor 17
|
|
chr9_+_133154896
|
1.991
|
NM_032728
|
PPAPDC3
|
phosphatidic acid phosphatase type 2 domain containing 3
|
|
chr2_+_233029076
|
1.944
|
NM_001631
|
ALPI
|
alkaline phosphatase, intestinal
|
|
chr19_-_13808101
|
1.924
|
|
LOC284454
|
hypothetical LOC284454
|
|
chr1_+_200429740
|
1.902
|
NM_001017403
|
LGR6
|
leucine-rich repeat containing G protein-coupled receptor 6
|
|
chr19_+_2200112
|
1.897
|
NM_000479
|
AMH
|
anti-Mullerian hormone
|
|
chr11_+_43920385
|
1.880
|
NM_001145033
|
C11orf96
|
chromosome 11 open reading frame 96
|
|
chr1_-_6402467
|
1.843
|
NM_019089
|
HES2
|
hairy and enhancer of split 2 (Drosophila)
|
|
chr7_+_2525922
|
1.782
|
NM_001040167
NM_001040168
|
LFNG
|
LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase
|
|
chr9_-_132804243
|
1.770
|
NM_001145106
|
FIBCD1
|
fibrinogen C domain containing 1
|
|
chr3_-_13032130
|
1.741
|
|
|
|
|
chr22_+_19248733
|
1.718
|
|
MED15
|
mediator complex subunit 15
|
|
chr9_-_139183675
|
1.717
|
|
|
|
|
chr4_+_8322358
|
1.688
|
NM_053044
|
HTRA3
|
HtrA serine peptidase 3
|
|
chr10_+_134060681
|
1.688
|
NM_001098637
NM_138499
|
PWWP2B
|
PWWP domain containing 2B
|
|
chr10_+_80498782
|
1.685
|
NM_020338
|
ZMIZ1
|
zinc finger, MIZ-type containing 1
|
|
chr16_+_2461500
|
1.645
|
NM_006181
|
NTN3
|
netrin 3
|
|
chr19_+_3473953
|
1.637
|
NM_001136197
NM_001136198
|
FZR1
|
fizzy/cell division cycle 20 related 1 (Drosophila)
|
|
chr1_-_226202210
|
1.636
|
NM_003395
|
WNT9A
|
wingless-type MMTV integration site family, member 9A
|
|
chr6_+_37245860
|
1.617
|
NM_002648
|
PIM1
|
pim-1 oncogene
|
|
chr19_+_55164668
|
1.602
|
|
SIGLEC16
|
sialic acid binding Ig-like lectin 16 (gene/pseudogene)
|
|
chr8_-_145085616
|
1.601
|
NM_201384
|
PLEC
|
plectin
|
|
chr1_+_1940605
|
1.584
|
NM_000815
|
GABRD
|
gamma-aminobutyric acid (GABA) A receptor, delta
|
|
chr5_+_176726918
|
1.579
|
|
RGS14
|
regulator of G-protein signaling 14
|
|
chr19_+_3045528
|
1.567
|
|
GNA11
|
guanine nucleotide binding protein (G protein), alpha 11 (Gq class)
|
|
chr11_-_75595098
|
1.534
|
NM_004626
|
WNT11
|
wingless-type MMTV integration site family, member 11
|
|
chr17_+_77453363
|
1.529
|
NM_148896
|
NPB
|
neuropeptide B
|
|
chr1_-_21868380
|
1.523
|
NM_001145657
NM_002885
|
RAP1GAP
|
RAP1 GTPase activating protein
|
|
chr7_+_100515439
|
1.506
|
NM_030961
|
TRIM56
|
tripartite motif containing 56
|
|
chr1_-_37991160
|
1.497
|
|
EPHA10
|
EPH receptor A10
|
|
chr8_+_145705253
|
1.491
|
NM_138431
|
MFSD3
|
major facilitator superfamily domain containing 3
|
|
chr10_+_135343649
|
1.482
|
NM_012147
|
DUX2
|
double homeobox 2
|
|
chr10_+_72826656
|
1.477
|
NM_001171930
NM_001171931
NM_001171932
NM_022124
NM_052836
|
CDH23
|
cadherin-related 23
|
|
chr16_+_711139
|
1.475
|
NM_023933
|
FAM173A
|
family with sequence similarity 173, member A
|
|
chr1_+_3378030
|
1.454
|
|
ARHGEF16
|
Rho guanine nucleotide exchange factor (GEF) 16
|
|
chr11_-_75599433
|
1.422
|
|
WNT11
|
wingless-type MMTV integration site family, member 11
|
|
chrX_-_106846253
|
1.411
|
NM_001015881
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr19_+_44266784
|
1.409
|
NM_001004318
|
PAPL
|
iron/zinc purple acid phosphatase-like protein
|
|
chr11_+_2422747
|
1.402
|
NM_000218
|
KCNQ1
|
potassium voltage-gated channel, KQT-like subfamily, member 1
|
|
chr12_+_108636569
|
1.401
|
NM_032829
|
C12orf34
|
chromosome 12 open reading frame 34
|
|
chr7_-_127833231
|
1.400
|
NM_001142573
NM_001142574
NM_001142575
|
IMPDH1
|
IMP (inosine 5'-monophosphate) dehydrogenase 1
|
|
chr4_+_8633116
|
1.394
|
NM_080819
|
GPR78
|
G protein-coupled receptor 78
|
|
chr11_+_74629597
|
1.392
|
NM_001195528
|
LOC441617
|
hypothetical LOC441617
|
|
chr22_+_20101661
|
1.391
|
NM_015094
|
HIC2
|
hypermethylated in cancer 2
|
|
chr8_-_140784416
|
1.391
|
NM_016601
|
KCNK9
|
potassium channel, subfamily K, member 9
|
|
chr2_+_94900904
|
1.383
|
NM_144705
|
TEKT4
|
tektin 4
|
|
chr1_+_1256585
|
1.382
|
NM_152228
|
TAS1R3
|
taste receptor, type 1, member 3
|
|
chr19_-_54857003
|
1.379
|
|
IRF3
|
interferon regulatory factor 3
|
|
chr21_+_42946719
|
1.379
|
NM_001001567
NM_001001568
NM_001001569
NM_001001570
NM_001001571
NM_001001572
NM_001001573
NM_001001574
NM_001001575
NM_001001576
NM_001001577
NM_001001578
NM_001001579
NM_001001580
NM_001001581
NM_001001582
NM_001001583
NM_001001584
NM_001001585
NM_002606
|
PDE9A
|
phosphodiesterase 9A
|
|
chr2_+_3620296
|
1.378
|
NM_024027
NM_199235
|
COLEC11
|
collectin sub-family member 11
|
|
chr11_-_66877519
|
1.378
|
NM_021173
|
POLD4
|
polymerase (DNA-directed), delta 4
|
|
chr16_+_84204424
|
1.360
|
NM_014615
|
KIAA0182
|
KIAA0182
|
|
chr4_+_3413489
|
1.335
|
NM_001528
|
HGFAC
|
HGF activator
|
|
chr20_-_61933012
|
1.334
|
|
ZBTB46
|
zinc finger and BTB domain containing 46
|
|
chr15_-_75693309
|
1.332
|
|
LINGO1
|
leucine rich repeat and Ig domain containing 1
|
|
chr16_-_1215254
|
1.326
|
NM_012467
|
TPSG1
|
tryptase gamma 1
|
|
chr21_+_44844924
|
1.323
|
NM_198689
|
KRTAP10-7
|
keratin associated protein 10-7
|
|
chr7_+_154943466
|
1.321
|
NM_001427
|
EN2
|
engrailed homeobox 2
|
|
chr22_-_20235372
|
1.318
|
NM_001128635
|
RIMBP3B
|
RIMS binding protein 3B
|
|
chr19_+_46390916
|
1.308
|
NM_030622
|
CYP2S1
|
cytochrome P450, family 2, subfamily S, polypeptide 1
|
|
chr20_+_61838479
|
1.307
|
|
LIME1
|
Lck interacting transmembrane adaptor 1
|
|
chr3_+_135996990
|
1.302
|
|
EPHB1
|
EPH receptor B1
|
|
chr19_-_59438411
|
1.300
|
NM_024318
|
LILRB3
LILRA6
|
leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 3
leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 6
|
|
chr1_-_1171964
|
1.297
|
NM_001014980
|
FAM132A
|
family with sequence similarity 132, member A
|
|
chr3_+_151608673
|
1.289
|
|
TSC22D2
|
TSC22 domain family, member 2
|
|
chr12_+_50981915
|
1.288
|
NM_002284
|
KRT86
|
keratin 86
|
|
chr11_-_66945190
|
1.281
|
|
PPP1CA
|
protein phosphatase 1, catalytic subunit, alpha isozyme
|
|
chr1_-_6474346
|
1.276
|
NM_020631
|
PLEKHG5
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5
|
|
chr8_-_145090892
|
1.275
|
NM_201381
|
PLEC
|
plectin
|
|
chr22_+_17539198
|
1.272
|
|
|
|
|
chr17_+_7269658
|
1.272
|
NM_175734
|
C17orf74
|
chromosome 17 open reading frame 74
|
|
chr10_+_135333667
|
1.267
|
NM_033178
NM_012147
NM_001127387
NM_001127388
NM_001127389
NM_001164467
NM_001127386
|
DUX4
DUX2
DUX4L7
DUX4L6
DUX4L5
DUX4L3
DUX4L2
|
double homeobox 4
double homeobox 2
double homeobox 4 like 7
double homeobox 4 like 6
double homeobox 4 like 5
double homeobox 4 like 3
double homeobox 4 like 2
|
|
chr6_-_168219380
|
1.257
|
NM_001122841
|
FRMD1
|
FERM domain containing 1
|
|
chr19_-_3928516
|
1.252
|
|
EEF2
|
eukaryotic translation elongation factor 2
|
|
chr19_+_46390980
|
1.246
|
|
CYP2S1
|
cytochrome P450, family 2, subfamily S, polypeptide 1
|
|
chr19_+_54883732
|
1.243
|
NM_001101340
|
C19orf76
|
chromosome 19 open reading frame 76
|
|
chr3_-_50515857
|
1.237
|
NM_001005505
NM_001174051
NM_006030
|
CACNA2D2
|
calcium channel, voltage-dependent, alpha 2/delta subunit 2
|
|
chr8_-_142307854
|
1.235
|
NM_001080431
|
SLC45A4
|
solute carrier family 45, member 4
|
|
chr22_-_19122047
|
1.232
|
|
SCARF2
|
scavenger receptor class F, member 2
|
|
chr1_-_6585268
|
1.228
|
|
KLHL21
|
kelch-like 21 (Drosophila)
|
|
chr4_+_3737872
|
1.227
|
|
ADRA2C
|
adrenergic, alpha-2C-, receptor
|
|
chr2_-_233060774
|
1.226
|
NM_004826
|
ECEL1
|
endothelin converting enzyme-like 1
|
|
chr12_+_106236281
|
1.225
|
NM_001018072
|
BTBD11
|
BTB (POZ) domain containing 11
|
|
chr1_-_155095289
|
1.223
|
NM_014215
|
INSRR
|
insulin receptor-related receptor
|
|
chr14_+_104117100
|
1.217
|
NM_001008404
|
C14orf180
|
chromosome 14 open reading frame 180
|
|
chr8_-_145099963
|
1.216
|
NM_201379
|
PLEC
|
plectin
|
|
chr9_+_130620540
|
1.208
|
NM_004435
|
ENDOG
|
endonuclease G
|
|
chr5_-_525916
|
1.201
|
|
LOC25845
|
hypothetical LOC25845
|
|
chr1_-_925332
|
1.197
|
NM_001142467
NM_021170
|
HES4
|
hairy and enhancer of split 4 (Drosophila)
|
|
chr1_+_1360765
|
1.189
|
NM_022834
NM_199121
|
VWA1
|
von Willebrand factor A domain containing 1
|
|
chr20_+_61838389
|
1.188
|
NM_017806
|
LIME1
|
Lck interacting transmembrane adaptor 1
|
|
chr16_-_11270619
|
1.185
|
NM_005425
|
TNP2
|
transition protein 2 (during histone to protamine replacement)
|
|
chr2_+_241156776
|
1.183
|
NM_018226
|
RNPEPL1
|
arginyl aminopeptidase (aminopeptidase B)-like 1
|
|
chr14_+_99859427
|
1.170
|
NM_207117
|
SLC25A47
|
solute carrier family 25, member 47
|
|
chr21_+_44569537
|
1.169
|
|
PFKL
|
phosphofructokinase, liver
|
|
chr1_-_1525309
|
1.166
|
|
LOC643988
|
hypothetical LOC643988
|
|
chr22_+_19248810
|
1.160
|
|
MED15
|
mediator complex subunit 15
|
|
chr19_-_1062972
|
1.154
|
|
SBNO2
|
strawberry notch homolog 2 (Drosophila)
|
|
chr7_-_158073121
|
1.153
|
NM_002847
NM_130842
NM_130843
|
PTPRN2
|
protein tyrosine phosphatase, receptor type, N polypeptide 2
|
|
chr19_+_3045266
|
1.141
|
NM_002067
|
GNA11
|
guanine nucleotide binding protein (G protein), alpha 11 (Gq class)
|
|
chr22_+_19649460
|
1.139
|
|
AIFM3
|
apoptosis-inducing factor, mitochondrion-associated, 3
|
|
chr7_+_73341640
|
1.134
|
NM_003388
NM_032421
|
CLIP2
|
CAP-GLY domain containing linker protein 2
|
|
chr9_-_139047112
|
1.129
|
NM_004479
|
FUT7
|
fucosyltransferase 7 (alpha (1,3) fucosyltransferase)
|
|
chr1_+_1099145
|
1.128
|
NM_001130045
|
TTLL10
|
tubulin tyrosine ligase-like family, member 10
|
|
chr4_+_191232653
|
1.127
|
NM_033178
NM_012147
NM_001177376
NM_001127387
NM_001127388
NM_001127389
NM_001164467
NM_001127386
|
DUX4
DUX2
DUX4L4
DUX4L7
DUX4L6
DUX4L5
DUX4L3
DUX4L2
|
double homeobox 4
double homeobox 2
double homeobox 4 like 4
double homeobox 4 like 7
double homeobox 4 like 6
double homeobox 4 like 5
double homeobox 4 like 3
double homeobox 4 like 2
|
|
chr17_+_75507750
|
1.124
|
|
|
|
|
chr10_+_88404285
|
1.114
|
NM_001030015
NM_033282
|
OPN4
|
opsin 4
|
|
chr4_+_191239247
|
1.114
|
NM_033178
NM_012147
NM_001177376
NM_001127388
NM_001127389
NM_001164467
NM_001127386
|
DUX4
DUX2
DUX4L4
DUX4L6
DUX4L5
DUX4L3
DUX4L2
|
double homeobox 4
double homeobox 2
double homeobox 4 like 4
double homeobox 4 like 6
double homeobox 4 like 5
double homeobox 4 like 3
double homeobox 4 like 2
|
|
chr8_+_145461356
|
1.109
|
NM_001008271
NM_001080514
|
SCXA
SCXB
|
scleraxis homolog A (mouse)
scleraxis homolog B (mouse)
|
|
chr9_-_128924782
|
1.094
|
|
ANGPTL2
|
angiopoietin-like 2
|
|
chr16_+_73810383
|
1.093
|
NM_001906
|
CTRB1
CTRB2
|
chymotrypsinogen B1
chymotrypsinogen B2
|
|
chr22_-_42619139
|
1.092
|
NM_001177675
NM_138814
|
PNPLA5
|
patatin-like phospholipase domain containing 5
|
|
chr22_-_19122107
|
1.091
|
NM_153334
NM_182895
|
SCARF2
|
scavenger receptor class F, member 2
|
|
chr19_+_482688
|
1.077
|
NM_004359
|
CDC34
|
cell division cycle 34 homolog (S. cerevisiae)
|
|
chr1_+_6226838
|
1.074
|
NM_001024598
|
HES3
|
hairy and enhancer of split 3 (Drosophila)
|
|
chr10_+_80740584
|
1.069
|
|
ZMIZ1
|
zinc finger, MIZ-type containing 1
|
|
chr22_+_49333327
|
1.060
|
NM_138433
|
KLHDC7B
|
kelch domain containing 7B
|
|
chr17_-_3447084
|
1.060
|
NM_018727
|
TRPV1
|
transient receptor potential cation channel, subfamily V, member 1
|
|
chr3_+_135996734
|
1.059
|
NM_004441
|
EPHB1
|
EPH receptor B1
|
|
chr1_+_1104939
|
1.057
|
NM_153254
|
TTLL10
|
tubulin tyrosine ligase-like family, member 10
|
|
chr19_+_55571493
|
1.055
|
NM_007121
|
NR1H2
|
nuclear receptor subfamily 1, group H, member 2
|
|
chr19_-_50355247
|
1.051
|
NM_198478
|
NKPD1
|
NTPase, KAP family P-loop domain containing 1
|
|
chr9_-_128924819
|
1.040
|
NM_012098
|
ANGPTL2
|
angiopoietin-like 2
|
|
chr21_+_43462209
|
1.033
|
NM_000394
|
CRYAA
|
crystallin, alpha A
|
|
chr3_-_10027770
|
1.031
|
NM_001008737
|
LOC401052
|
hypothetical LOC401052
|
|
chr22_-_30928497
|
1.031
|
NM_001159546
|
RFPL2
|
ret finger protein-like 2
|
|
chrX_-_55037235
|
1.028
|
NM_002625
|
PFKFB1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1
|
|
chr1_+_152806874
|
1.014
|
NM_000748
|
CHRNB2
|
cholinergic receptor, nicotinic, beta 2 (neuronal)
|
|
chr10_+_135330357
|
1.012
|
NM_033178
NM_012147
NM_001127387
NM_001127388
NM_001127389
NM_001164467
NM_001127386
|
DUX4
DUX2
DUX4L7
DUX4L6
DUX4L5
DUX4L3
DUX4L2
|
double homeobox 4
double homeobox 2
double homeobox 4 like 7
double homeobox 4 like 6
double homeobox 4 like 5
double homeobox 4 like 3
double homeobox 4 like 2
|
|
chr19_-_2378874
|
1.008
|
NM_012458
|
TIMM13
|
translocase of inner mitochondrial membrane 13 homolog (yeast)
|
|
chr1_+_154878498
|
1.007
|
|
BCAN
|
brevican
|
|
chr11_-_785005
|
1.006
|
|
SLC25A22
|
solute carrier family 25 (mitochondrial carrier: glutamate), member 22
|
|
chr20_-_61639104
|
1.004
|
NM_005975
|
PTK6
|
PTK6 protein tyrosine kinase 6
|
|
chr8_-_41624030
|
1.004
|
NM_152568
|
NKX6-3
|
NK6 homeobox 3
|
|
chr17_-_36346240
|
1.003
|
|
KRT23
|
keratin 23 (histone deacetylase inducible)
|
|
chr17_-_29930431
|
1.002
|
NM_207454
|
C17orf102
|
chromosome 17 open reading frame 102
|
|
chr19_-_3922032
|
1.000
|
|
DAPK3
|
death-associated protein kinase 3
|
|
chr22_-_15982142
|
1.000
|
NM_031890
NM_001163079
|
CECR6
|
cat eye syndrome chromosome region, candidate 6
|
|
chr7_+_45163891
|
1.000
|
NM_005856
|
RAMP3
|
receptor (G protein-coupled) activity modifying protein 3
|
|
chr1_-_39871226
|
0.993
|
|
HEYL
|
hairy/enhancer-of-split related with YRPW motif-like
|
|
chr17_+_14145051
|
0.989
|
NM_006041
|
HS3ST3B1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 3B1
|
|
chr16_-_790699
|
0.988
|
NM_016541
|
GNG13
|
guanine nucleotide binding protein (G protein), gamma 13
|
|
chr12_+_130878870
|
0.987
|
NM_016155
|
MMP17
|
matrix metallopeptidase 17 (membrane-inserted)
|
|
chr1_-_1840564
|
0.979
|
NM_178545
|
TMEM52
|
transmembrane protein 52
|
|
chr16_+_2226468
|
0.979
|
NM_001374
|
DNASE1L2
|
deoxyribonuclease I-like 2
|
|
chr19_+_482737
|
0.979
|
|
CDC34
|
cell division cycle 34 homolog (S. cerevisiae)
|
|
chr11_+_65100074
|
0.977
|
NM_001099409
|
EHBP1L1
|
EH domain binding protein 1-like 1
|
|
chr19_-_8118367
|
0.976
|
NM_032447
|
FBN3
|
fibrillin 3
|
|
chr4_+_191235959
|
0.973
|
NM_001177376
|
DUX4L4
|
double homeobox 4 like 4
|
|
chr7_+_128293699
|
0.971
|
NM_001195150
|
LOC100130705
|
hypothetical protein LOC100130705
|
|
chr9_+_139255559
|
0.966
|
|
TUBB2C
|
tubulin, beta 2C
|
|
chr15_-_66285427
|
0.964
|
NM_001031733
NM_033429
|
CALML4
|
calmodulin-like 4
|
|
chr1_+_152806924
|
0.959
|
|
CHRNB2
|
cholinergic receptor, nicotinic, beta 2 (neuronal)
|
|
chr11_+_67567022
|
0.959
|
NM_006053
|
TCIRG1
|
T-cell, immune regulator 1, ATPase, H+ transporting, lysosomal V0 subunit A3
|
|
chr1_-_20122638
|
0.958
|
NM_014589
|
PLA2G2E
|
phospholipase A2, group IIE
|
|
chr19_+_18543613
|
0.958
|
NM_001033930
NM_003333
|
UBA52
|
ubiquitin A-52 residue ribosomal protein fusion product 1
|
|
chr18_+_75724655
|
0.955
|
NM_012283
|
KCNG2
|
potassium voltage-gated channel, subfamily G, member 2
|
|
chr9_-_128924706
|
0.955
|
|
ANGPTL2
|
angiopoietin-like 2
|
|
chr1_-_6243621
|
0.954
|
NM_207370
|
GPR153
|
G protein-coupled receptor 153
|
|
chr3_-_195553297
|
0.942
|
NM_001080513
|
CPN2
|
carboxypeptidase N, polypeptide 2
|
|
chr3_-_48516486
|
0.940
|
NM_016479
|
SHISA5
|
shisa homolog 5 (Xenopus laevis)
|
|
chr19_-_3922089
|
0.935
|
|
DAPK3
|
death-associated protein kinase 3
|
|
chr9_-_138958889
|
0.927
|
|
FBXW5
|
F-box and WD repeat domain containing 5
|
|
chr20_+_1254033
|
0.925
|
|
LOC100507495
|
hypothetical LOC100507495
|
|
chr1_+_148788410
|
0.924
|
NM_019032
NM_025008
|
ADAMTSL4
|
ADAMTS-like 4
|
|
chr11_-_118693021
|
0.921
|
NM_006500
|
MCAM
|
melanoma cell adhesion molecule
|
|
chr6_+_108593907
|
0.907
|
NM_003269
|
NR2E1
|
nuclear receptor subfamily 2, group E, member 1
|
|
chr19_-_3931499
|
0.907
|
|
EEF2
|
eukaryotic translation elongation factor 2
|
|
chr7_-_127458231
|
0.907
|
NM_022143
|
LRRC4
|
leucine rich repeat containing 4
|
|
chr2_+_232165818
|
0.906
|
NM_152614
|
C2orf57
|
chromosome 2 open reading frame 57
|
|
chr1_+_44643513
|
0.904
|
NM_018150
|
RNF220
|
ring finger protein 220
|
|
chrX_+_67784235
|
0.903
|
NM_001142503
NM_014725
|
STARD8
|
StAR-related lipid transfer (START) domain containing 8
|
|
chr6_+_31538934
|
0.902
|
NM_006674
|
HCP5
|
HLA complex P5
|
|
chr20_-_61108831
|
0.900
|
NM_080606
|
BHLHE23
|
basic helix-loop-helix family, member e23
|
|
chr19_+_6415259
|
0.900
|
NM_139161
NM_174881
|
CRB3
|
crumbs homolog 3 (Drosophila)
|
|
chr8_-_145097031
|
0.897
|
NM_201380
|
PLEC
|
plectin
|
|
chr16_-_87460514
|
0.894
|
NM_001080487
|
PABPN1L
|
poly(A) binding protein, nuclear 1-like (cytoplasmic)
|
|
chr4_+_191226073
|
0.893
|
NM_012147
NM_001177376
NM_001127388
NM_001127389
NM_001164467
|
DUX2
DUX4L4
DUX4L6
DUX4L5
DUX4L3
|
double homeobox 2
double homeobox 4 like 4
double homeobox 4 like 6
double homeobox 4 like 5
double homeobox 4 like 3
|
|
chr16_+_2138608
|
0.893
|
NM_014353
|
RAB26
|
RAB26, member RAS oncogene family
|
|
chr11_-_46364681
|
0.886
|
NM_000741
|
CHRM4
|
cholinergic receptor, muscarinic 4
|
|
chr20_-_21442663
|
0.884
|
NM_002509
|
NKX2-2
|
NK2 homeobox 2
|
|
chr9_+_139255519
|
0.883
|
NM_006088
|
TUBB2C
|
tubulin, beta 2C
|
|
chr10_+_71908535
|
0.882
|
NM_014431
|
KIAA1274
|
KIAA1274
|
|
chr9_-_135213149
|
0.881
|
|
SURF1
|
surfeit 1
|
|
chr17_-_77462506
|
0.880
|
NM_001184917
NM_002861
|
PCYT2
|
phosphate cytidylyltransferase 2, ethanolamine
|