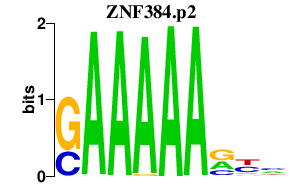
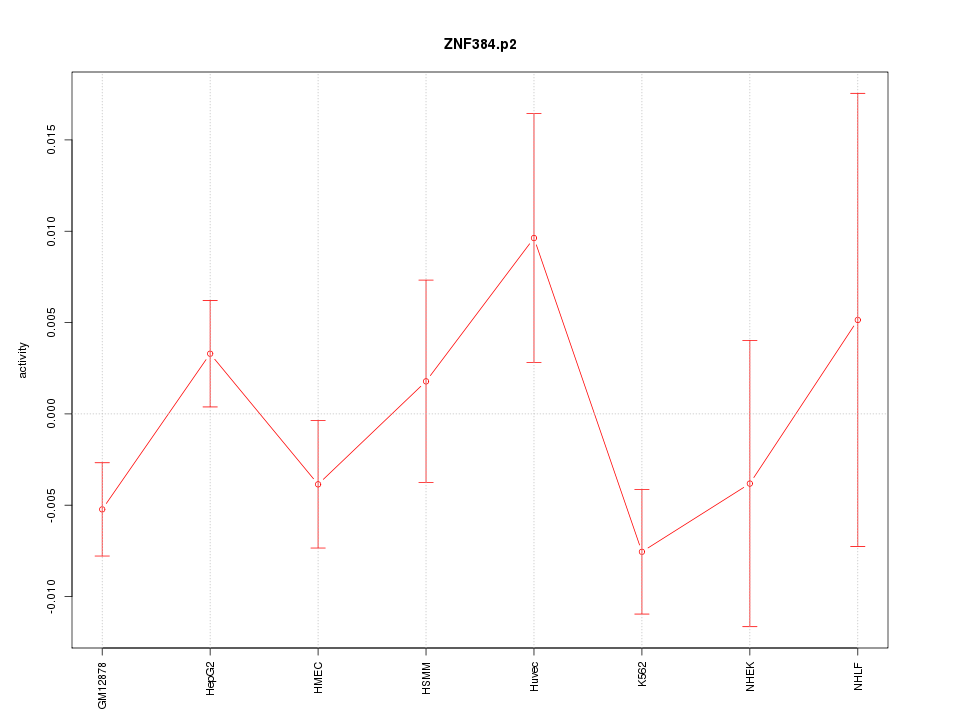
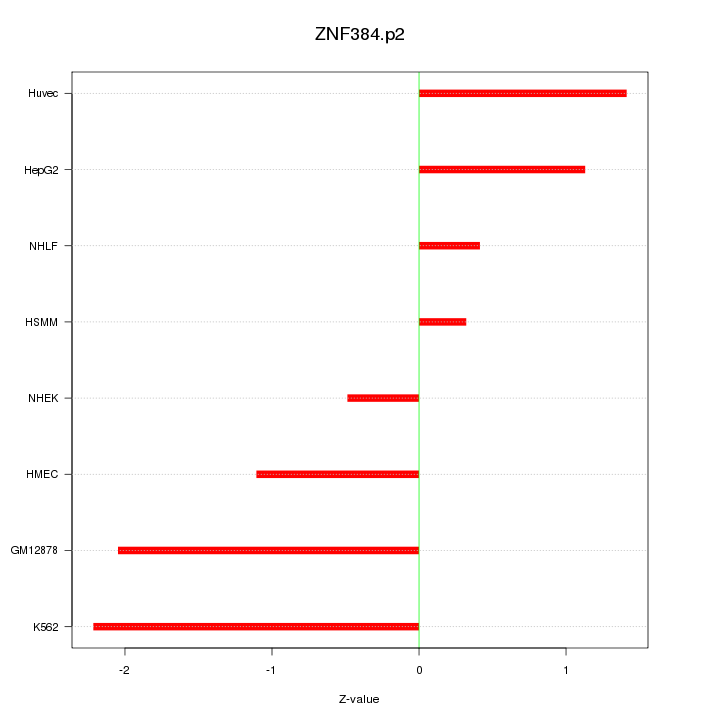
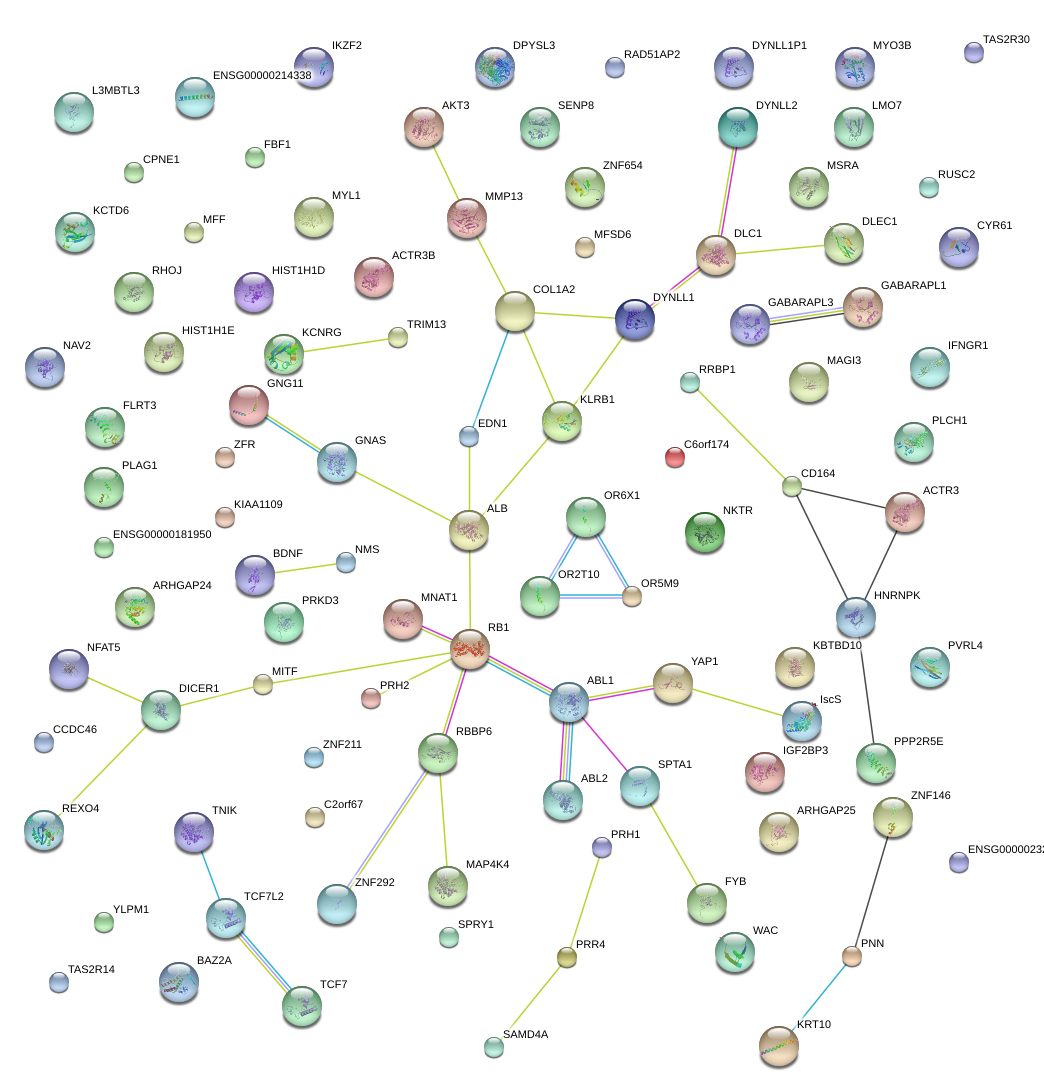
|
chr11_+_101488380
|
5.821
|
NM_001195045
|
YAP1
|
Yes-associated protein 1
|
|
chr3_+_88270951
|
4.761
|
NM_018293
|
ZNF654
|
zinc finger protein 654
|
|
chr8_-_13018123
|
4.194
|
NM_001164271
|
DLC1
|
deleted in liver cancer 1
|
|
chr6_-_26343139
|
3.084
|
NM_005320
|
HIST1H1D
|
histone cluster 1, H1d
|
|
chr11_-_55987452
|
2.562
|
NM_001004743
|
OR5M9
|
olfactory receptor, family 5, subfamily M, member 9
|
|
chr13_+_75232797
|
2.530
|
NM_015842
|
LMO7
|
LIM domain 7
|
|
chr11_-_102331643
|
2.354
|
NM_002427
|
MMP13
|
matrix metallopeptidase 13 (collagenase 3)
|
|
chr11_-_27637771
|
2.099
|
NM_001143816
NM_170735
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr16_+_68284252
|
2.006
|
|
NFAT5
|
nuclear factor of activated T-cells 5, tonicity-responsive
|
|
chr16_+_24475129
|
1.973
|
|
RBBP6
|
retinoblastoma binding protein 6
|
|
chr1_-_242073094
|
1.967
|
NM_005465
NM_181690
|
AKT3
|
v-akt murine thymoma viral oncogene homolog 3 (protein kinase B, gamma)
|
|
chr14_+_38719462
|
1.941
|
|
PNN
|
pinin, desmosome associated protein
|
|
chr6_+_88026338
|
1.896
|
|
ZNF292
|
zinc finger protein 292
|
|
chr2_-_37397669
|
1.883
|
NM_005813
|
PRKD3
|
protein kinase D3
|
|
chr13_+_49487390
|
1.879
|
NM_173605
NM_199464
|
TRIM13
KCNRG
|
tripartite motif containing 13
potassium channel regulator
|
|
chr6_+_12398514
|
1.803
|
NM_001168319
NM_001955
|
EDN1
|
endothelin 1
|
|
chr6_-_109795925
|
1.784
|
|
CD164
|
CD164 molecule, sialomucin
|
|
chr5_-_146813343
|
1.779
|
NM_001387
|
DPYSL3
|
dihydropyrimidinase-like 3
|
|
chr9_-_85773899
|
1.773
|
|
HNRNPK
|
heterogeneous nuclear ribonucleoprotein K
|
|
chr10_+_114699953
|
1.770
|
NM_001146274
NM_001146283
NM_001146284
NM_001146285
NM_001146286
NM_001198525
NM_001198526
NM_001198527
NM_001198528
NM_001198529
NM_001198530
NM_001198531
NM_030756
|
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box)
|
|
chr12_-_11178109
|
1.768
|
NM_001097643
|
TAS2R30
|
taste receptor, type 2, member 30
|
|
chr13_+_47948586
|
1.767
|
|
RB1
|
retinoblastoma 1
|
|
chr22_+_29696662
|
1.767
|
|
|
|
|
chr14_+_74353417
|
1.765
|
|
YLPM1
|
YLP motif containing 1
|
|
chr8_+_9990471
|
1.754
|
NM_001135671
|
MSRA
|
methionine sulfoxide reductase A
|
|
chr7_+_93388951
|
1.740
|
NM_004126
|
GNG11
|
guanine nucleotide binding protein (G protein), gamma 11
|
|
chr20_-_17589122
|
1.697
|
|
RRBP1
|
ribosome binding protein 1 homolog 180kDa (dog)
|
|
chr4_+_87070449
|
1.692
|
NM_031305
|
ARHGAP24
|
Rho GTPase activating protein 24
|
|
chr2_+_170742900
|
1.662
|
NM_001083615
NM_001171642
NM_138995
|
MYO3B
|
myosin IIIB
|
|
chr2_+_227900471
|
1.637
|
NM_020194
|
MFF
|
mitochondrial fission factor
|
|
chr20_-_14266200
|
1.594
|
NM_013281
NM_198391
|
FLRT3
|
fibronectin leucine rich transmembrane protein 3
|
|
chr20_+_56903595
|
1.591
|
|
GNAS
|
GNAS complex locus
|
|
chr5_-_39238818
|
1.526
|
|
FYB
|
FYN binding protein
|
|
chr10_+_28918672
|
1.519
|
|
WAC
|
WW domain containing adaptor with coiled-coil
|
|
chr2_+_114390975
|
1.519
|
|
ACTR3
|
ARP3 actin-related protein 3 homolog (yeast)
|
|
chr1_-_156923111
|
1.509
|
NM_003126
|
SPTA1
|
spectrin, alpha, erythrocytic 1 (elliptocytosis 2)
|
|
chr19_+_41418723
|
1.499
|
|
ZNF146
|
zinc finger protein 146
|
|
chr1_-_58920315
|
1.494
|
|
|
|
|
chr2_-_213723298
|
1.487
|
NM_016260
|
IKZF2
|
IKAROS family zinc finger 2 (Helios)
|
|
chr2_-_17563186
|
1.468
|
NM_001099218
|
RAD51AP2
|
RAD51 associated protein 2
|
|
chr7_+_93862137
|
1.465
|
|
COL1A2
PLAG1
|
collagen, type I, alpha 2
pleiomorphic adenoma gene 1
|
|
chr6_-_127882192
|
1.450
|
NM_001012279
|
C6orf174
|
chromosome 6 open reading frame 174
|
|
chr1_-_246823691
|
1.418
|
NM_001004693
|
OR2T10
|
olfactory receptor, family 2, subfamily T, member 10
|
|
chr15_+_22781292
|
1.412
|
|
PAR5
|
Prader-Willi/Angelman syndrome-5
|
|
chr14_+_60271203
|
1.410
|
NM_001177963
NM_002431
|
MNAT1
|
menage a trois homolog 1, cyclin H assembly factor (Xenopus laevis)
|
|
chr3_-_61703850
|
1.377
|
|
|
|
|
chr11_-_123130435
|
1.372
|
NM_001005188
|
OR6X1
|
olfactory receptor, family 6, subfamily X, member 1
|
|
chr5_-_32439067
|
1.371
|
|
ZFR
|
zinc finger RNA binding protein
|
|
chr12_-_55295628
|
1.360
|
|
BAZ2A
|
bromodomain adjacent to zinc finger domain, 2A
|
|
chr2_+_170074457
|
1.358
|
NM_006063
|
KBTBD10
|
kelch repeat and BTB (POZ) domain containing 10
|
|
chr19_+_62844143
|
1.340
|
|
ZNF211
|
zinc finger protein 211
|
|
chr11_+_20027185
|
1.340
|
|
NAV2
|
neuron navigator 2
|
|
chr14_+_54104383
|
1.337
|
NM_001161576
NM_015589
|
SAMD4A
|
sterile alpha motif domain containing 4A
|
|
chr14_-_63076155
|
1.326
|
|
PPP2R5E
|
protein phosphatase 2, regulatory subunit B', epsilon isoform
|
|
chr17_-_61253122
|
1.324
|
NM_001037325
|
CCDC46
|
coiled-coil domain containing 46
|
|
chr17_-_36232366
|
1.301
|
NM_000421
|
KRT10
|
keratin 10
|
|
chr9_+_132579088
|
1.297
|
NM_007313
|
ABL1
|
c-abl oncogene 1, non-receptor tyrosine kinase
|
|
chr2_-_210888010
|
1.274
|
NM_079420
|
MYL1
|
myosin, light chain 1, alkali; skeletal, fast
|
|
chr4_+_74500748
|
1.263
|
|
ALB
|
albumin
|
|
chr14_-_94669547
|
1.260
|
NM_001195573
|
DICER1
|
dicer 1, ribonuclease type III
|
|
chr19_+_41419165
|
1.246
|
|
ZNF146
|
zinc finger protein 146
|
|
chr3_+_42617109
|
1.237
|
NM_005385
|
NKTR
|
natural killer-tumor recognition sequence
|
|
chr10_+_114700775
|
1.231
|
|
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box)
|
|
chr3_+_70068440
|
1.218
|
NM_000248
NM_001184968
NM_198158
NM_198178
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr12_-_11215396
|
1.215
|
NM_001098538
NM_006250
|
PRH1-PRR4
PRR4
TAS2R14
PRH1
|
PRH1-PRR4 read-through transcript
proline rich 4 (lacrimal)
taste receptor, type 2, member 14
proline-rich protein HaeIII subfamily 1
|
|
chr12_+_10256682
|
1.205
|
NM_031412
|
GABARAPL1
|
GABA(A) receptor-associated protein like 1
|
|
chr15_+_70193652
|
1.202
|
NM_001166340
|
SENP8
|
SUMO/sentrin specific peptidase family member 8
|
|
chr3_-_156876798
|
1.184
|
NM_001130960
NM_001130961
|
PLCH1
|
phospholipase C, eta 1
|
|
chr3_+_58459131
|
1.175
|
NM_153331
|
KCTD6
|
potassium channel tetramerisation domain containing 6
|
|
chr7_-_23476497
|
1.161
|
NM_006547
|
IGF2BP3
|
insulin-like growth factor 2 mRNA binding protein 3
|
|
chr1_+_169747788
|
1.152
|
|
PRRC2C
|
proline-rich coiled-coil 2C
|
|
chr1_+_85819004
|
1.145
|
NM_001554
|
CYR61
|
cysteine-rich, angiogenic inducer, 61
|
|
chr6_+_130381408
|
1.142
|
NM_001007102
NM_032438
|
L3MBTL3
|
l(3)mbt-like 3 (Drosophila)
|
|
chr2_-_210726539
|
1.135
|
|
C2orf67
|
chromosome 2 open reading frame 67
|
|
chr5_-_121842680
|
1.134
|
|
|
|
|
chr2_+_68815416
|
1.122
|
NM_001007231
NM_001166277
|
ARHGAP25
|
Rho GTPase activating protein 25
|
|
chr2_+_100453375
|
1.118
|
NM_001011717
|
NMS
|
neuromedin S
|
|
chr2_+_191009265
|
1.118
|
|
MFSD6
|
major facilitator superfamily domain containing 6
|
|
chr14_+_62740878
|
1.108
|
NM_020663
|
RHOJ
|
ras homolog gene family, member J
|
|
chr1_+_113734997
|
1.104
|
NM_001142782
NM_152900
|
MAGI3
|
membrane associated guanylate kinase, WW and PDZ domain containing 3
|
|
chr4_+_124540114
|
1.088
|
NM_005841
|
SPRY1
|
sprouty homolog 1, antagonist of FGF signaling (Drosophila)
|
|
chr4_+_123380569
|
1.079
|
|
KIAA1109
|
KIAA1109
|
|
chr20_-_33705244
|
1.065
|
NM_003915
|
CPNE1
|
copine I
|
|
chr9_+_35528890
|
1.058
|
|
RUSC2
|
RUN and SH3 domain containing 2
|
|
chr2_+_101680594
|
1.058
|
NM_004834
NM_145686
NM_145687
|
MAP4K4
|
mitogen-activated protein kinase kinase kinase kinase 4
|
|
chr1_+_143807876
|
1.050
|
|
SEC22B
|
SEC22 vesicle trafficking protein homolog B (S. cerevisiae) (gene/pseudogene)
|
|
chr17_+_26446082
|
1.047
|
|
NF1
|
neurofibromin 1
|
|
chr16_+_55373241
|
1.039
|
|
NUP93
|
nucleoporin 93kDa
|
|
chr7_+_17349425
|
1.037
|
|
AHR
|
aryl hydrocarbon receptor
|
|
chr10_-_35403173
|
1.037
|
NM_001198778
NM_001198779
|
CUL2
|
cullin 2
|
|
chr2_+_189547358
|
1.030
|
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr9_+_35528628
|
1.026
|
NM_001135999
|
RUSC2
|
RUN and SH3 domain containing 2
|
|
chr12_+_54106304
|
1.015
|
NM_001005183
|
OR6C76
|
olfactory receptor, family 6, subfamily C, member 76
|
|
chr6_+_151688327
|
1.014
|
NM_144497
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chr13_-_85271329
|
1.004
|
NM_032229
|
SLITRK6
|
SLIT and NTRK-like family, member 6
|
|
chr12_+_53927248
|
1.004
|
NM_001005490
|
OR6C74
|
olfactory receptor, family 6, subfamily C, member 74
|
|
chr15_+_97010219
|
1.004
|
NM_000875
|
IGF1R
|
insulin-like growth factor 1 receptor
|
|
chr5_-_58918031
|
1.003
|
NM_006203
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr3_+_153499883
|
1.002
|
NM_207293
NM_207294
NM_207295
NM_207296
NM_207297
|
MBNL1
|
muscleblind-like (Drosophila)
|
|
chr7_+_117651947
|
0.988
|
NM_019644
|
ANKRD7
|
ankyrin repeat domain 7
|
|
chr5_+_67620007
|
0.986
|
NM_181524
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha)
|
|
chr2_+_134728299
|
0.986
|
NM_002410
|
MGAT5
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase
|
|
chr3_+_148610516
|
0.983
|
|
ZIC1
|
Zic family member 1 (odd-paired homolog, Drosophila)
|
|
chr6_+_88024152
|
0.979
|
|
ZNF292
|
zinc finger protein 292
|
|
chr20_+_9236446
|
0.978
|
NM_000933
|
PLCB4
|
phospholipase C, beta 4
|
|
chr20_+_41519947
|
0.977
|
|
SRSF6
|
serine/arginine-rich splicing factor 6
|
|
chr1_-_156800017
|
0.977
|
NM_001160325
|
OR6P1
|
olfactory receptor, family 6, subfamily P, member 1
|
|
chr10_+_128583984
|
0.970
|
NM_001380
|
DOCK1
|
dedicator of cytokinesis 1
|
|
chr11_-_100505667
|
0.949
|
NM_000926
|
PGR
|
progesterone receptor
|
|
chr18_+_3443771
|
0.947
|
NM_173211
|
TGIF1
|
TGFB-induced factor homeobox 1
|
|
chr6_-_48144344
|
0.946
|
NM_001013732
NM_207499
|
C6orf138
|
chromosome 6 open reading frame 138
|
|
chr19_+_37528340
|
0.938
|
NM_001136156
NM_014910
|
ZNF507
|
zinc finger protein 507
|
|
chr1_+_162795328
|
0.936
|
NM_002585
|
PBX1
|
pre-B-cell leukemia homeobox 1
|
|
chr2_+_189547323
|
0.930
|
NM_000090
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr4_-_149577404
|
0.925
|
|
|
|
|
chr17_+_72692889
|
0.920
|
NM_001144001
|
SEC14L1
|
SEC14-like 1 (S. cerevisiae)
|
|
chr9_-_21375258
|
0.919
|
NM_000605
|
IFNA2
|
interferon, alpha 2
|
|
chr18_-_38949481
|
0.916
|
NM_002930
|
RIT2
|
Ras-like without CAAX 2
|
|
chr18_-_28247186
|
0.913
|
|
|
|
|
chr15_-_53996597
|
0.912
|
NM_198400
|
NEDD4
|
neural precursor cell expressed, developmentally down-regulated 4
|
|
chr16_+_68156497
|
0.911
|
NM_001113178
NM_006599
NM_138713
NM_138714
NM_173214
|
NFAT5
|
nuclear factor of activated T-cells 5, tonicity-responsive
|
|
chr21_+_25856311
|
0.909
|
|
MIR155HG
|
MIR155 host gene (non-protein coding)
|
|
chr6_+_45404031
|
0.906
|
NM_001015051
NM_001024630
|
RUNX2
|
runt-related transcription factor 2
|
|
chr1_-_103346639
|
0.905
|
NM_001190709
NM_001854
NM_080629
NM_080630
|
COL11A1
|
collagen, type XI, alpha 1
|
|
chr17_+_29606408
|
0.898
|
NM_002982
|
CCL2
|
chemokine (C-C motif) ligand 2
|
|
chr8_+_87947788
|
0.897
|
NM_173538
|
CNBD1
|
cyclic nucleotide binding domain containing 1
|
|
chr15_+_22778233
|
0.897
|
|
SNRPN
|
small nuclear ribonucleoprotein polypeptide N
|
|
chr9_-_124280025
|
0.895
|
NM_001004451
|
OR1J1
|
olfactory receptor, family 1, subfamily J, member 1
|
|
chr7_-_93027057
|
0.894
|
NM_001164738
|
CALCR
|
calcitonin receptor
|
|
chr14_+_38720085
|
0.893
|
|
PNN
|
pinin, desmosome associated protein
|
|
chr15_-_77994652
|
0.893
|
NM_001100879
|
ST20
|
suppressor of tumorigenicity 20
|
|
chr21_-_26345294
|
0.892
|
|
|
|
|
chr2_+_166034402
|
0.890
|
NM_001172173
|
CSRNP3
|
cysteine-serine-rich nuclear protein 3
|
|
chr8_+_50147455
|
0.886
|
NM_001007176
|
C8orf22
|
chromosome 8 open reading frame 22
|
|
chr13_-_101852028
|
0.882
|
NM_175929
|
FGF14
|
fibroblast growth factor 14
|
|
chr20_+_41519882
|
0.880
|
NM_006275
|
SRSF6
|
serine/arginine-rich splicing factor 6
|
|
chr20_-_33783409
|
0.879
|
|
RBM39
|
RNA binding motif protein 39
|
|
chr3_+_120496180
|
0.864
|
|
ARHGAP31
|
Rho GTPase activating protein 31
|
|
chr11_-_7804094
|
0.863
|
NM_153445
|
OR5P3
|
olfactory receptor, family 5, subfamily P, member 3
|
|
chr5_+_41940216
|
0.850
|
NM_175921
|
C5orf51
|
chromosome 5 open reading frame 51
|
|
chr9_-_106420305
|
0.847
|
NM_001001956
|
OR13C9
|
olfactory receptor, family 13, subfamily C, member 9
|
|
chr21_-_14840506
|
0.846
|
NM_022136
|
SAMSN1
|
SAM domain, SH3 domain and nuclear localization signals 1
|
|
chr1_+_157167965
|
0.841
|
NM_152501
NM_198928
NM_198929
NM_198930
|
PYHIN1
|
pyrin and HIN domain family, member 1
|
|
chr9_+_124477135
|
0.841
|
NM_001005234
|
OR1L3
|
olfactory receptor, family 1, subfamily L, member 3
|
|
chr3_+_101910823
|
0.841
|
NM_001007565
NM_001195479
NM_006070
|
TFG
|
TRK-fused gene
|
|
chr9_+_107249897
|
0.837
|
NM_001145313
NM_031919
NM_207647
|
FSD1L
|
fibronectin type III and SPRY domain containing 1-like
|
|
chr1_+_224317062
|
0.835
|
|
H3F3A
LOC440926
|
H3 histone, family 3A
H3 histone, family 3A pseudogene
|
|
chr6_+_121798443
|
0.834
|
NM_000165
|
GJA1
|
gap junction protein, alpha 1, 43kDa
|
|
chr12_+_99391870
|
0.833
|
|
NR1H4
|
nuclear receptor subfamily 1, group H, member 4
|
|
chr8_-_17797127
|
0.832
|
NM_147203
NM_201553
NM_004467
NM_201552
|
FGL1
|
fibrinogen-like 1
|
|
chr10_-_61570665
|
0.831
|
NM_001149
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr21_-_31107431
|
0.829
|
NM_175857
|
KRTAP8-1
|
keratin associated protein 8-1
|
|
chr11_+_16716751
|
0.828
|
|
C11orf58
|
chromosome 11 open reading frame 58
|
|
chr14_+_31868229
|
0.827
|
NM_004274
|
AKAP6
|
A kinase (PRKA) anchor protein 6
|
|
chr12_+_73219826
|
0.824
|
|
|
|
|
chr4_+_38740246
|
0.824
|
NM_015990
NM_199039
|
KLHL5
|
kelch-like 5 (Drosophila)
|
|
chr4_+_95595419
|
0.813
|
|
PDLIM5
|
PDZ and LIM domain 5
|
|
chr8_-_39814860
|
0.808
|
NM_001464
|
ADAM2
|
ADAM metallopeptidase domain 2
|
|
chr7_+_80069439
|
0.805
|
NM_001001547
|
CD36
|
CD36 molecule (thrombospondin receptor)
|
|
chr12_+_20739555
|
0.803
|
NM_001145945
NM_001145946
NM_001145944
NM_017435
|
SLCO1C1
|
solute carrier organic anion transporter family, member 1C1
|
|
chr1_+_178232379
|
0.802
|
|
CEP350
|
centrosomal protein 350kDa
|
|
chr8_-_101384633
|
0.801
|
NM_015435
|
RNF19A
|
ring finger protein 19A
|
|
chr7_+_134226727
|
0.799
|
|
CALD1
|
caldesmon 1
|
|
chr9_-_21340885
|
0.795
|
NM_021002
|
IFNA6
|
interferon, alpha 6
|
|
chr3_+_101911051
|
0.795
|
NM_001195478
|
TFG
|
TRK-fused gene
|
|
chr4_-_77163609
|
0.793
|
NM_001565
|
CXCL10
|
chemokine (C-X-C motif) ligand 10
|
|
chr5_+_121325554
|
0.792
|
NM_152546
|
SRFBP1
|
serum response factor binding protein 1
|
|
chr9_+_71009746
|
0.789
|
NM_001170415
NM_001170416
|
TJP2
|
tight junction protein 2 (zona occludens 2)
|
|
chr4_-_74154272
|
0.786
|
NM_173827
|
COX18
|
COX18 cytochrome c oxidase assembly homolog (S. cerevisiae)
|
|
chr18_+_64616296
|
0.784
|
NM_024781
|
CCDC102B
|
coiled-coil domain containing 102B
|
|
chr1_-_21288259
|
0.783
|
|
EIF4G3
|
eukaryotic translation initiation factor 4 gamma, 3
|
|
chr20_+_2031418
|
0.771
|
|
STK35
|
serine/threonine kinase 35
|
|
chr2_+_33213111
|
0.770
|
NM_000627
NM_001166264
NM_001166265
NM_001166266
|
LTBP1
|
latent transforming growth factor beta binding protein 1
|
|
chr19_-_9186512
|
0.769
|
NM_001005191
|
OR7D4
|
olfactory receptor, family 7, subfamily D, member 4
|
|
chr3_-_116272911
|
0.767
|
NM_001164343
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr3_-_52269467
|
0.766
|
|
WDR82
|
WD repeat domain 82
|
|
chr5_+_57914658
|
0.761
|
NM_138453
|
RAB3C
|
RAB3C, member RAS oncogene family
|
|
chr10_+_102748062
|
0.760
|
|
LZTS2
|
leucine zipper, putative tumor suppressor 2
|
|
chr9_-_106705867
|
0.760
|
|
ABCA1
|
ATP-binding cassette, sub-family A (ABC1), member 1
|
|
chr20_-_2031521
|
0.760
|
|
|
|
|
chr13_+_31503436
|
0.756
|
NM_023037
|
FRY
|
furry homolog (Drosophila)
|
|
chr12_-_60872814
|
0.755
|
NM_178539
|
FAM19A2
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A2
|
|
chr11_+_58967192
|
0.752
|
NM_001004728
|
OR5A1
|
olfactory receptor, family 5, subfamily A, member 1
|
|
chr9_-_114814286
|
0.751
|
NM_001101338
|
ZNF883
|
zinc finger protein 883
|
|
chr3_+_138159396
|
0.749
|
NM_144717
|
IL20RB
|
interleukin 20 receptor beta
|
|
chr20_+_13924263
|
0.747
|
|
MACROD2
|
MACRO domain containing 2
|
|
chr2_-_151826504
|
0.745
|
NM_198557
|
RBM43
|
RNA binding motif protein 43
|
|
chr8_-_103206310
|
0.741
|
NM_001040630
|
NCALD
|
neurocalcin delta
|
|
chr12_-_23995109
|
0.739
|
|
SOX5
|
SRY (sex determining region Y)-box 5
|
|
chr1_+_190871868
|
0.737
|
NM_002927
NM_144766
|
RGS13
|
regulator of G-protein signaling 13
|
|
chr7_+_149842850
|
0.737
|
NM_153236
|
GIMAP7
|
GTPase, IMAP family member 7
|
|
chr7_+_134226690
|
0.732
|
NM_033139
NM_033140
|
CALD1
|
caldesmon 1
|
|
chr9_-_106401514
|
0.731
|
NM_001004482
|
OR13C5
|
olfactory receptor, family 13, subfamily C, member 5
|
|
chr4_+_53975597
|
0.728
|
|
FIP1L1
|
FIP1 like 1 (S. cerevisiae)
|
|
chr11_-_55815113
|
0.728
|
NM_001005199
|
OR8H1
|
olfactory receptor, family 8, subfamily H, member 1
|
|
chr15_-_54175954
|
0.727
|
|
RFX7
|
regulatory factor X, 7
|
|
chr3_-_187124290
|
0.726
|
|
TRA2B
|
transformer 2 beta homolog (Drosophila)
|
|
chr1_-_71285847
|
0.724
|
NM_001126044
NM_198714
NM_198715
NM_198716
NM_198717
NM_198718
NM_198719
|
PTGER3
|
prostaglandin E receptor 3 (subtype EP3)
|
|
chr4_+_111616630
|
0.722
|
NM_001977
|
ENPEP
|
glutamyl aminopeptidase (aminopeptidase A)
|
|
chr4_+_77575276
|
0.721
|
NM_020859
|
SHROOM3
|
shroom family member 3
|