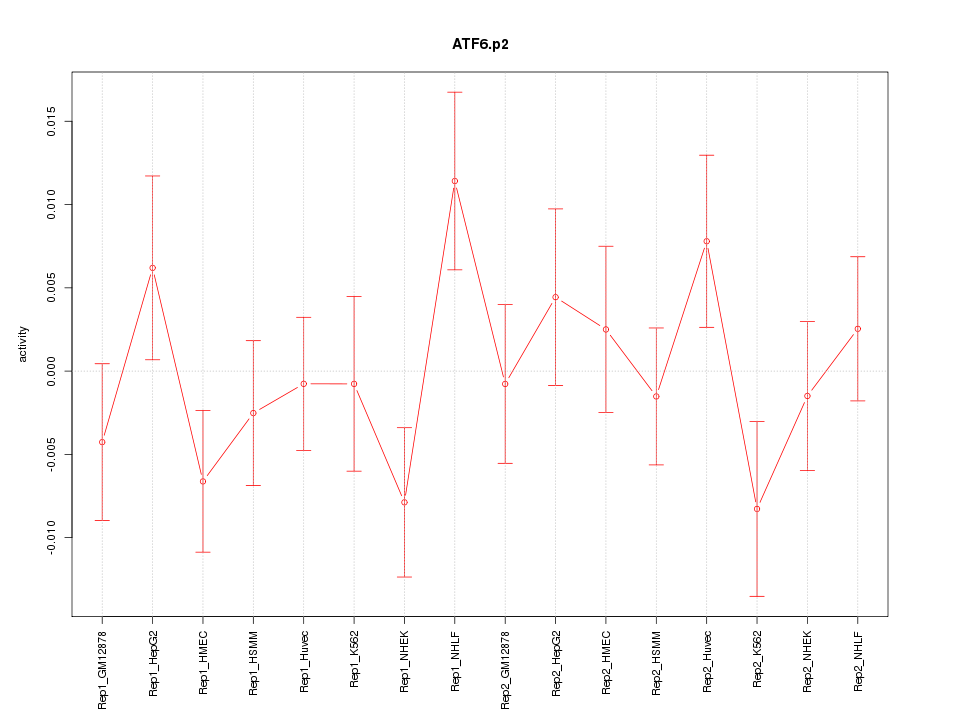
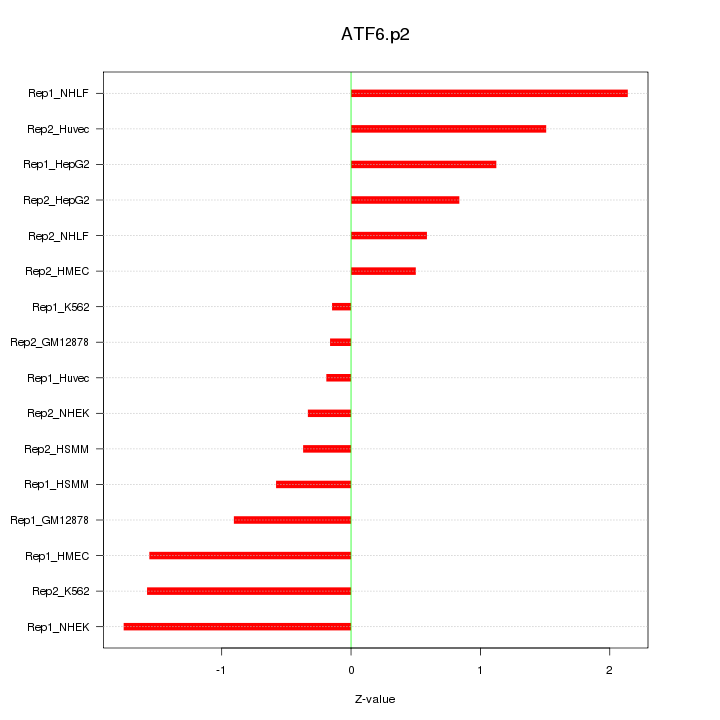
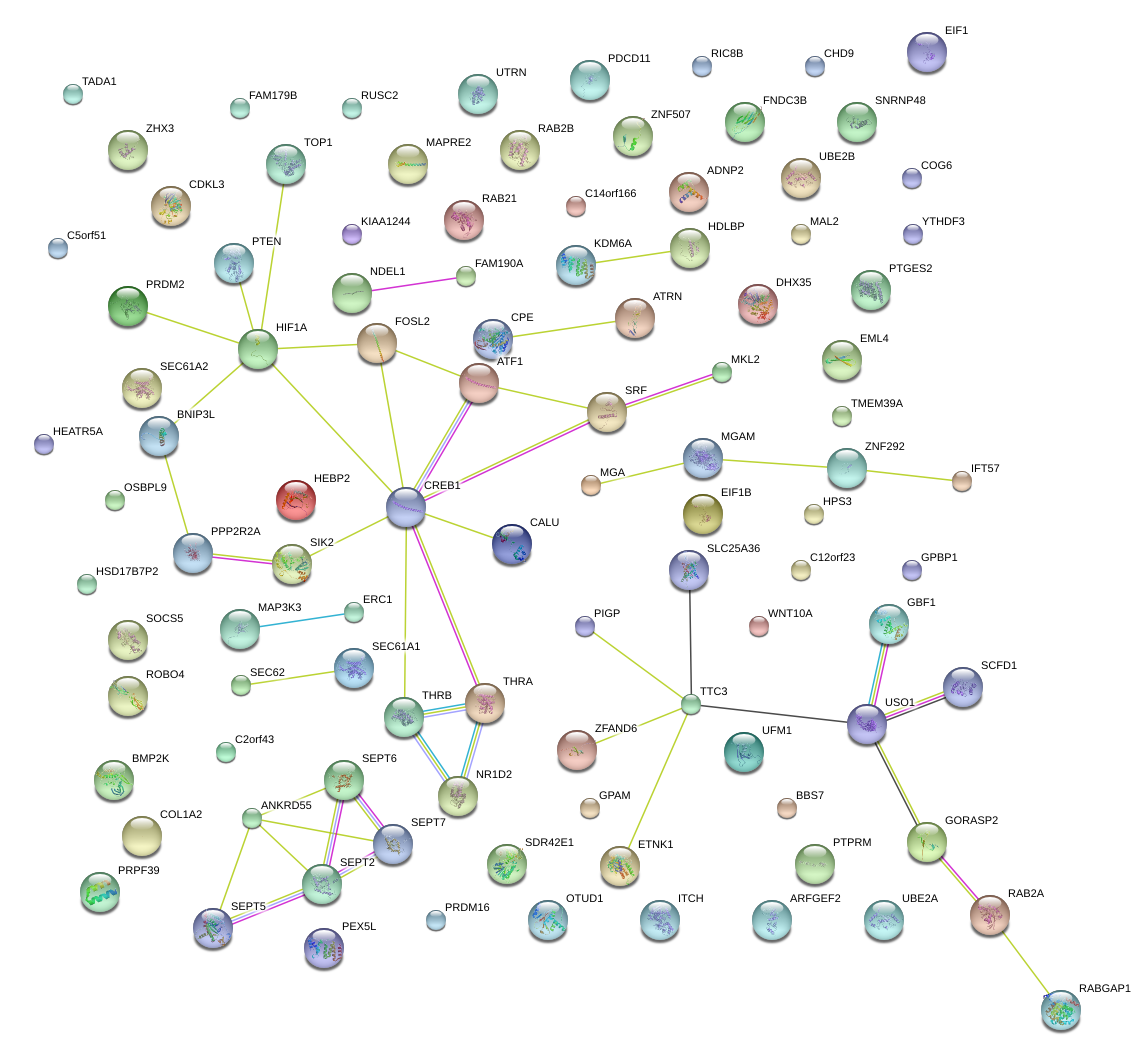
|
chr2_+_46779553
|
1.639
|
NM_014011
|
SOCS5
|
suppressor of cytokine signaling 5
|
|
chr8_+_26296439
|
1.618
|
NM_004331
|
BNIP3L
|
BCL2/adenovirus E1B 19kDa interacting protein 3-like
|
|
chr12_+_70434927
|
1.479
|
|
RAB21
|
RAB21, member RAS oncogene family
|
|
chr16_+_51722285
|
1.422
|
|
CHD9
|
chromodomain helicase DNA binding protein 9
|
|
chr2_+_46779827
|
1.396
|
NM_144949
|
SOCS5
|
suppressor of cytokine signaling 5
|
|
chr21_+_37367432
|
1.353
|
NM_001001894
|
TTC3
|
tetratricopeptide repeat domain 3
|
|
chr12_+_70434920
|
1.325
|
NM_014999
|
RAB21
|
RAB21, member RAS oncogene family
|
|
chr12_+_70435176
|
1.288
|
|
RAB21
|
RAB21, member RAS oncogene family
|
|
chr3_+_171167243
|
1.182
|
NM_003262
|
SEC62
|
SEC62 homolog (S. cerevisiae)
|
|
chr20_+_46971618
|
1.154
|
NM_006420
|
ARFGEF2
|
ADP-ribosylation factor guanine nucleotide-exchange factor 2 (brefeldin A-inhibited)
|
|
chr21_-_37367259
|
1.118
|
NM_153682
|
PIGP
|
phosphatidylinositol glycan anchor biosynthesis, class P
|
|
chr12_+_970613
|
1.022
|
NM_178039
NM_178040
|
ERC1
|
ELKS/RAB6-interacting/CAST family member 1
|
|
chr2_+_208103068
|
1.015
|
|
CREB1
|
cAMP responsive element binding protein 1
|
|
chr6_+_144649142
|
0.978
|
|
UTRN
|
utrophin
|
|
chr5_+_41940216
|
0.925
|
NM_175921
|
C5orf51
|
chromosome 5 open reading frame 51
|
|
chr12_+_105873803
|
0.910
|
|
C12orf23
|
chromosome 12 open reading frame 23
|
|
chr18_+_30875297
|
0.899
|
NM_014268
|
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2
|
|
chr18_+_7557313
|
0.882
|
NM_001105244
NM_002845
|
PTPRM
|
protein tyrosine phosphatase, receptor type, M
|
|
chr13_+_37821964
|
0.875
|
|
UFM1
|
ubiquitin-fold modifier 1
|
|
chr3_-_120665113
|
0.867
|
NM_018266
|
TMEM39A
|
transmembrane protein 39A
|
|
chr20_+_37024383
|
0.859
|
NM_001190809
NM_021931
|
DHX35
|
DEAH (Asp-Glu-Ala-His) box polypeptide 35
|
|
chr8_+_64243662
|
0.837
|
NM_152758
|
YTHDF3
|
YTH domain family, member 3
|
|
chr8_+_64243741
|
0.834
|
|
YTHDF3
|
YTH domain family, member 3
|
|
chr2_-_241860923
|
0.832
|
NM_203346
|
HDLBP
|
high density lipoprotein binding protein
|
|
chr21_-_37366684
|
0.828
|
NM_153681
|
PIGP
|
phosphatidylinositol glycan anchor biosynthesis, class P
|
|
chr2_+_208103001
|
0.822
|
|
CREB1
|
cAMP responsive element binding protein 1
|
|
chr3_+_150330060
|
0.800
|
NM_032383
|
HPS3
|
Hermansky-Pudlak syndrome 3
|
|
chr6_+_7535404
|
0.795
|
NM_152551
|
SNRNP48
|
small nuclear ribonucleoprotein 48kDa (U11/U12)
|
|
chr7_+_93861808
|
0.780
|
NM_000089
|
COL1A2
|
collagen, type I, alpha 2
|
|
chr15_+_39739864
|
0.780
|
NM_001080541
NM_001164273
|
MGA
|
MAX gene associated
|
|
chr2_-_241860917
|
0.768
|
|
HDLBP
|
high density lipoprotein binding protein
|
|
chr12_+_105873669
|
0.766
|
NM_152261
|
C12orf23
|
chromosome 12 open reading frame 23
|
|
chr9_+_124742932
|
0.757
|
NM_012197
|
RABGAP1
|
RAB GTPase activating protein 1
|
|
chr2_+_28469199
|
0.749
|
|
FOSL2
|
FOS-like antigen 2
|
|
chr2_+_28469275
|
0.744
|
NM_005253
|
FOSL2
|
FOS-like antigen 2
|
|
chr19_+_37528340
|
0.744
|
NM_001136156
NM_014910
|
ZNF507
|
zinc finger protein 507
|
|
chr2_-_241860871
|
0.739
|
|
HDLBP
|
high density lipoprotein binding protein
|
|
chr8_+_120289735
|
0.738
|
NM_052886
|
MAL2
|
mal, T-cell differentiation protein 2 (gene/pseudogene)
|
|
chr15_+_39739933
|
0.711
|
|
MGA
|
MAX gene associated
|
|
chr10_+_89613347
|
0.708
|
|
PTEN
|
phosphatase and tensin homolog
|
|
chr2_+_28469172
|
0.703
|
|
FOSL2
|
FOS-like antigen 2
|
|
chr2_+_208102937
|
0.700
|
|
CREB1
|
cAMP responsive element binding protein 1
|
|
chr3_-_181237210
|
0.699
|
NM_016559
|
PEX5L
|
peroxisomal biogenesis factor 5-like
|
|
chr14_+_61231795
|
0.690
|
NM_001530
NM_181054
|
HIF1A
|
hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor)
|
|
chr2_+_42249896
|
0.663
|
NM_001145076
NM_019063
|
EML4
|
echinoderm microtubule associated protein like 4
|
|
chr14_-_30927931
|
0.655
|
NM_015473
|
HEATR5A
|
HEAT repeat containing 5A
|
|
chr5_+_133734768
|
0.653
|
NM_003337
|
UBE2B
|
ubiquitin-conjugating enzyme E2B (RAD6 homolog)
|
|
chr10_+_89613399
|
0.632
|
|
PTEN
|
phosphatase and tensin homolog
|
|
chr14_+_30161285
|
0.630
|
|
SCFD1
|
sec1 family domain containing 1
|
|
chr9_+_35528890
|
0.624
|
|
RUSC2
|
RUN and SH3 domain containing 2
|
|
chr4_+_166519392
|
0.619
|
NM_001873
|
CPE
|
carboxypeptidase E
|
|
chr14_+_44501083
|
0.604
|
NM_015091
|
FAM179B
|
family with sequence similarity 179, member B
|
|
chr14_+_30161267
|
0.602
|
NM_016106
NM_182835
|
SCFD1
|
sec1 family domain containing 1
|
|
chr20_+_39090875
|
0.599
|
NM_003286
|
TOP1
|
topoisomerase (DNA) I
|
|
chr14_+_44622916
|
0.596
|
NM_017922
|
PRPF39
|
PRP39 pre-mRNA processing factor 39 homolog (S. cerevisiae)
|
|
chr20_-_10148137
|
0.587
|
|
LOC100131208
|
similar to hCG2045825
|
|
chr11_+_110978309
|
0.582
|
NM_015191
|
SIK2
|
salt-inducible kinase 2
|
|
chr3_+_173240983
|
0.573
|
NM_001135095
|
FNDC3B
|
fibronectin type III domain containing 3B
|
|
chr13_+_37821941
|
0.572
|
NM_016617
|
UFM1
|
ubiquitin-fold modifier 1
|
|
chr5_-_133734616
|
0.572
|
|
CDKL3
|
cyclin-dependent kinase-like 3
|
|
chr13_+_39127758
|
0.571
|
NM_001145079
NM_020751
|
COG6
|
component of oligomeric golgi complex 6
|
|
chr10_-_113933457
|
0.569
|
NM_020918
|
GPAM
|
glycerol-3-phosphate acyltransferase, mitochondrial
|
|
chr3_+_142143329
|
0.567
|
NM_001104647
NM_018155
|
SLC25A36
|
solute carrier family 25, member 36
|
|
chr2_+_171494026
|
0.565
|
|
GORASP2
|
golgi reassembly stacking protein 2, 55kDa
|
|
chr16_-_80602546
|
0.562
|
NM_145168
|
SDR42E1
|
short chain dehydrogenase/reductase family 42E, member 1
|
|
chr20_+_39091026
|
0.560
|
|
TOP1
|
topoisomerase (DNA) I
|
|
chr10_-_89613173
|
0.556
|
NM_001126049
|
KLLN
|
killin, p53-regulated DNA replication inhibitor
|
|
chr4_+_76868763
|
0.553
|
|
USO1
|
USO1 vesicle docking protein homolog (yeast)
|
|
chr3_-_24511456
|
0.548
|
|
THRB
|
thyroid hormone receptor, beta (erythroblastic leukemia viral (v-erb-a) oncogene homolog 2, avian)
|
|
chr17_+_59053518
|
0.543
|
NM_002401
NM_203351
|
MAP3K3
|
mitogen-activated protein kinase kinase kinase 3
|
|
chr10_+_23768203
|
0.534
|
NM_001145373
|
OTUD1
|
OTU domain containing 1
|
|
chr20_+_3399615
|
0.529
|
NM_139321
NM_139322
|
ATRN
|
attractin
|
|
chr3_-_109423858
|
0.529
|
NM_018010
|
IFT57
|
intraflagellar transport 57 homolog (Chlamydomonas)
|
|
chr12_+_105692466
|
0.525
|
NM_018157
|
RIC8B
|
resistance to inhibitors of cholinesterase 8 homolog B (C. elegans)
|
|
chr5_-_99898788
|
0.525
|
|
|
|
|
chr2_+_208102860
|
0.524
|
NM_004379
NM_134442
|
CREB1
|
cAMP responsive element binding protein 1
|
|
chr6_+_87921980
|
0.512
|
NM_015021
|
ZNF292
|
zinc finger protein 292
|
|
chr2_+_171493870
|
0.507
|
NM_015530
|
GORASP2
|
golgi reassembly stacking protein 2, 55kDa
|
|
chr8_+_61592097
|
0.502
|
|
RAB2A
|
RAB2A, member RAS oncogene family
|
|
chr14_+_51525942
|
0.500
|
NM_016039
|
C14orf166
|
chromosome 14 open reading frame 166
|
|
chr5_+_56505412
|
0.500
|
NM_022913
|
GPBP1
|
GC-rich promoter binding protein 1
|
|
chr4_-_76868581
|
0.497
|
|
|
|
|
chr9_+_35528628
|
0.495
|
NM_001135999
|
RUSC2
|
RUN and SH3 domain containing 2
|
|
chr20_+_3399727
|
0.494
|
|
ATRN
|
attractin
|
|
chr8_+_61592105
|
0.493
|
NM_002865
|
RAB2A
|
RAB2A, member RAS oncogene family
|
|
chr3_+_23962518
|
0.489
|
NM_001145425
|
NR1D2
|
nuclear receptor subfamily 1, group D, member 2
|
|
chr17_+_59053515
|
0.489
|
|
MAP3K3
|
mitogen-activated protein kinase kinase kinase 3
|
|
chr4_+_79916576
|
0.488
|
|
BMP2K
|
BMP2 inducible kinase
|
|
chr4_+_79916555
|
0.487
|
NM_017593
NM_198892
|
BMP2K
|
BMP2 inducible kinase
|
|
chr8_+_61592048
|
0.484
|
|
RAB2A
|
RAB2A, member RAS oncogene family
|
|
chr14_+_51526071
|
0.484
|
|
C14orf166
|
chromosome 14 open reading frame 166
|
|
chr1_+_13899272
|
0.479
|
NM_001135610
|
PRDM2
|
PR domain containing 2, with ZNF domain
|
|
chr1_+_51855343
|
0.479
|
|
OSBPL9
|
oxysterol binding protein-like 9
|
|
chr4_-_123010918
|
0.476
|
NM_018190
NM_176824
|
BBS7
|
Bardet-Biedl syndrome 7
|
|
chr18_+_75967902
|
0.474
|
NM_014913
|
ADNP2
|
ADNP homeobox 2
|
|
chr2_+_219453487
|
0.471
|
NM_025216
|
WNT10A
|
wingless-type MMTV integration site family, member 10A
|
|
chr7_+_128166581
|
0.471
|
NM_001130674
NM_001219
|
CALU
|
calumenin
|
|
chr20_+_32414715
|
0.470
|
NM_031483
|
ITCH
|
itchy E3 ubiquitin protein ligase homolog (mouse)
|
|
chr8_+_61592278
|
0.467
|
|
RAB2A
|
RAB2A, member RAS oncogene family
|
|
chr8_+_26204960
|
0.465
|
|
PPP2R2A
|
protein phosphatase 2, regulatory subunit B, alpha
|
|
chr10_+_103995248
|
0.460
|
|
GBF1
|
golgi brefeldin A resistant guanine nucleotide exchange factor 1
|
|
chr17_+_8279860
|
0.459
|
NM_001025579
NM_030808
|
NDEL1
|
nudE nuclear distribution gene E homolog (A. nidulans)-like 1
|
|
chr4_+_91267706
|
0.456
|
NM_001145065
|
FAM190A
|
family with sequence similarity 190, member A
|
|
chr12_+_22669352
|
0.453
|
|
ETNK1
|
ethanolamine kinase 1
|
|
chr10_+_103995297
|
0.452
|
NM_004193
|
GBF1
|
golgi brefeldin A resistant guanine nucleotide exchange factor 1
|
|
chr5_+_56505608
|
0.451
|
|
GPBP1
|
GC-rich promoter binding protein 1
|
|
chr6_+_138524717
|
0.450
|
NM_020340
|
KIAA1244
|
KIAA1244
|
|
chr4_+_76868830
|
0.447
|
|
USO1
|
USO1 vesicle docking protein homolog (yeast)
|
|
chr2_-_20886293
|
0.445
|
NM_021925
|
C2orf43
|
chromosome 2 open reading frame 43
|
|
chr15_+_78139098
|
0.444
|
|
ZFAND6
|
zinc finger, AN1-type domain 6
|
|
chrX_+_44617346
|
0.444
|
NM_021140
|
KDM6A
|
lysine (K)-specific demethylase 6A
|
|
chr4_+_76868850
|
0.443
|
NM_003715
|
USO1
|
USO1 vesicle docking protein homolog (yeast)
|
|
chr20_-_39379533
|
0.440
|
|
ZHX3
|
zinc fingers and homeoboxes 3
|
|
chr16_+_14201687
|
0.440
|
|
MKL2
|
MKL/myocardin-like 2
|
|
chr3_+_129254032
|
0.440
|
|
|
|
|
chr2_+_241903395
|
0.439
|
NM_001008491
NM_006155
|
SEPT2
|
septin 2
|
|
chr15_+_78139073
|
0.438
|
NM_019006
|
ZFAND6
|
zinc finger, AN1-type domain 6
|
|
chr17_+_59053447
|
0.438
|
|
MAP3K3
|
mitogen-activated protein kinase kinase kinase 3
|
|
chr1_-_165112093
|
0.437
|
|
TADA1
|
transcriptional adaptor 1
|
|
chr3_+_129253997
|
0.436
|
|
SEC61A1
|
Sec61 alpha 1 subunit (S. cerevisiae)
|
|
chr3_+_129253816
|
0.435
|
NM_013336
|
SEC61A1
|
Sec61 alpha 1 subunit (S. cerevisiae)
|
|
chr3_+_40326254
|
0.434
|
|
EIF1B
|
eukaryotic translation initiation factor 1B
|
|
chr1_+_40399642
|
0.433
|
|
RLF
|
rearranged L-myc fusion
|
|
chr10_+_89613071
|
0.431
|
NM_000314
|
PTEN
|
phosphatase and tensin homolog
|
|
chr2_+_15649405
|
0.430
|
|
DDX1
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 1
|
|
chr4_+_170778265
|
0.430
|
NM_001829
NM_173872
|
CLCN3
|
chloride channel 3
|
|
chr6_+_124166767
|
0.427
|
NM_001040214
NM_153355
|
NKAIN2
|
Na+/K+ transporting ATPase interacting 2
|
|
chr1_+_40399618
|
0.426
|
NM_012421
|
RLF
|
rearranged L-myc fusion
|
|
chr22_-_37214480
|
0.426
|
NM_001098505
NM_030881
|
DDX17
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 17
|
|
chr7_+_128166631
|
0.425
|
|
CALU
|
calumenin
|
|
chr3_+_171558188
|
0.416
|
|
SKIL
|
SKI-like oncogene
|
|
chr20_+_5839973
|
0.414
|
NM_001819
|
CHGB
|
chromogranin B (secretogranin 1)
|
|
chr12_+_110935612
|
0.411
|
|
|
|
|
chr3_+_171558215
|
0.411
|
|
SKIL
|
SKI-like oncogene
|
|
chr7_+_39956396
|
0.410
|
|
|
|
|
chr3_+_12573513
|
0.409
|
NM_014160
|
MKRN2
|
makorin ring finger protein 2
|
|
chr3_+_143925545
|
0.408
|
|
|
|
|
chr5_-_34020445
|
0.406
|
NM_001012509
NM_016180
|
SLC45A2
|
solute carrier family 45, member 2
|
|
chr12_+_21546012
|
0.405
|
|
GOLT1B
|
golgi transport 1B
|
|
chr3_+_129253977
|
0.405
|
|
SEC61A1
|
Sec61 alpha 1 subunit (S. cerevisiae)
|
|
chr5_+_99898922
|
0.401
|
NM_198507
|
FAM174A
|
family with sequence similarity 174, member A
|
|
chr7_+_128166636
|
0.398
|
|
CALU
|
calumenin
|
|
chr4_+_79916489
|
0.398
|
|
BMP2K
|
BMP2 inducible kinase
|
|
chr6_-_127822227
|
0.398
|
NM_014702
|
KIAA0408
|
KIAA0408
|
|
chr14_-_38642165
|
0.397
|
NM_006364
|
SEC23A
|
Sec23 homolog A (S. cerevisiae)
|
|
chr5_+_149845503
|
0.396
|
|
NDST1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1
|
|
chr14_-_38642029
|
0.395
|
|
SEC23A
|
Sec23 homolog A (S. cerevisiae)
|
|
chr3_+_171558143
|
0.395
|
NM_001145098
NM_005414
|
SKIL
|
SKI-like oncogene
|
|
chr1_-_181871473
|
0.394
|
|
ARPC5
|
actin related protein 2/3 complex, subunit 5, 16kDa
|
|
chr7_+_64136078
|
0.394
|
|
CCT6P3
|
chaperonin containing TCP1, subunit 6 (zeta) pseudogene 3
|
|
chr10_+_49184658
|
0.392
|
|
MAPK8
|
mitogen-activated protein kinase 8
|
|
chr14_-_34253473
|
0.392
|
NM_138638
|
CFL2
|
cofilin 2 (muscle)
|
|
chr3_+_40326197
|
0.391
|
|
EIF1B
|
eukaryotic translation initiation factor 1B
|
|
chr3_+_40326176
|
0.387
|
NM_005875
|
EIF1B
|
eukaryotic translation initiation factor 1B
|
|
chr2_+_177785667
|
0.387
|
NM_194247
|
HNRNPA3P1
HNRNPA3
|
heterogeneous nuclear ribonucleoprotein A3 pseudogene 1
heterogeneous nuclear ribonucleoprotein A3
|
|
chr9_+_113433364
|
0.387
|
NM_001015882
NM_004125
|
DNAJC25
DNAJC25-GNG10
|
DnaJ (Hsp40) homolog, subfamily C , member 25
DNAJC25-GNG10 readthrough
|
|
chr1_+_152459918
|
0.387
|
NM_014847
|
UBAP2L
|
ubiquitin associated protein 2-like
|
|
chr2_-_236741352
|
0.386
|
NM_001485
|
GBX2
|
gastrulation brain homeobox 2
|
|
chr11_+_107384819
|
0.385
|
|
CUL5
|
cullin 5
|
|
chrX_+_21302317
|
0.385
|
NM_001168647
NM_001168648
NM_001168649
NM_014927
|
CNKSR2
|
connector enhancer of kinase suppressor of Ras 2
|
|
chr2_+_86521999
|
0.385
|
NM_001146688
|
KDM3A
|
lysine (K)-specific demethylase 3A
|
|
chr7_+_128166664
|
0.385
|
|
CALU
|
calumenin
|
|
chr8_-_38245694
|
0.384
|
NM_001102559
NM_001102560
NM_032483
|
PPAPDC1B
|
phosphatidic acid phosphatase type 2 domain containing 1B
|
|
chr3_+_142939827
|
0.384
|
|
RNF7
|
ring finger protein 7
|
|
chr3_+_101462375
|
0.383
|
NM_018309
|
TBC1D23
|
TBC1 domain family, member 23
|
|
chr2_-_241903657
|
0.382
|
|
HDLBP
|
high density lipoprotein binding protein
|
|
chr12_-_10657397
|
0.379
|
NM_018048
|
MAGOHB
|
mago-nashi homolog B (Drosophila)
|
|
chr10_-_38305458
|
0.375
|
NM_145011
|
ZNF25
|
zinc finger protein 25
|
|
chr12_+_122635006
|
0.373
|
NM_006815
|
TMED2
|
transmembrane emp24 domain trafficking protein 2
|
|
chr15_-_29950070
|
0.371
|
|
OTUD7A
|
OTU domain containing 7A
|
|
chr8_+_9450800
|
0.371
|
|
TNKS
|
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase
|
|
chr7_+_39956480
|
0.369
|
NM_003718
NM_031267
|
CDK13
|
cyclin-dependent kinase 13
|
|
chr12_+_110935644
|
0.368
|
|
ERP29
|
endoplasmic reticulum protein 29
|
|
chr3_+_150191884
|
0.368
|
NM_001184720
NM_001184721
NM_004130
|
GYG1
|
glycogenin 1
|
|
chr12_+_122635033
|
0.368
|
|
TMED2
|
transmembrane emp24 domain trafficking protein 2
|
|
chr15_-_29950188
|
0.368
|
|
OTUD7A
|
OTU domain containing 7A
|
|
chr12_+_22669275
|
0.367
|
NM_001039481
NM_018638
|
ETNK1
|
ethanolamine kinase 1
|
|
chr3_+_143925904
|
0.367
|
NM_003304
|
TRPC1
|
transient receptor potential cation channel, subfamily C, member 1
|
|
chr7_+_39956633
|
0.364
|
|
CDK13
|
cyclin-dependent kinase 13
|
|
chr8_+_9450849
|
0.362
|
NM_003747
|
TNKS
|
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase
|
|
chr15_+_78139018
|
0.361
|
|
ZFAND6
|
zinc finger, AN1-type domain 6
|
|
chr11_+_12652427
|
0.360
|
NM_021961
|
TEAD1
|
TEA domain family member 1 (SV40 transcriptional enhancer factor)
|
|
chr3_+_173240126
|
0.360
|
|
FNDC3B
|
fibronectin type III domain containing 3B
|
|
chr10_-_111673191
|
0.359
|
|
XPNPEP1
|
X-prolyl aminopeptidase (aminopeptidase P) 1, soluble
|
|
chr1_-_165112157
|
0.357
|
NM_053053
|
TADA1
|
transcriptional adaptor 1
|
|
chr5_+_108112537
|
0.356
|
|
FER
|
fer (fps/fes related) tyrosine kinase
|
|
chr6_-_128883125
|
0.356
|
NM_002844
NM_001135648
|
PTPRK
|
protein tyrosine phosphatase, receptor type, K
|
|
chr12_+_70120028
|
0.355
|
NM_003667
|
LGR5
|
leucine-rich repeat containing G protein-coupled receptor 5
|
|
chr20_+_34523555
|
0.352
|
NM_183006
|
DLGAP4
|
discs, large (Drosophila) homolog-associated protein 4
|
|
chr3_+_33815064
|
0.349
|
NM_001162429
NM_013374
|
PDCD6IP
|
programmed cell death 6 interacting protein
|
|
chr1_+_52294554
|
0.349
|
|
BTF3L4
|
basic transcription factor 3-like 4
|
|
chr2_+_30223253
|
0.348
|
NM_001127399
NM_001127400
NM_001127401
NM_016061
|
YPEL5
|
yippee-like 5 (Drosophila)
|
|
chr14_-_64040072
|
0.347
|
NM_006977
|
ZBTB25
|
zinc finger and BTB domain containing 25
|
|
chr7_+_93862137
|
0.347
|
|
COL1A2
PLAG1
|
collagen, type I, alpha 2
pleiomorphic adenoma gene 1
|
|
chr11_+_82545784
|
0.347
|
NM_015885
|
PCF11
|
PCF11, cleavage and polyadenylation factor subunit, homolog (S. cerevisiae)
|
|
chr14_-_44500871
|
0.345
|
NM_017658
|
KLHL28
|
kelch-like 28 (Drosophila)
|
|
chr10_-_81576162
|
0.344
|
|
|
|
|
chr3_-_24511180
|
0.343
|
NM_001128176
NM_000461
NM_001128177
|
THRB
|
thyroid hormone receptor, beta (erythroblastic leukemia viral (v-erb-a) oncogene homolog 2, avian)
|
|
chr7_+_128166688
|
0.342
|
|
CALU
|
calumenin
|
|
chr11_+_7965442
|
0.342
|
NM_003754
|
EIF3F
|
eukaryotic translation initiation factor 3, subunit F
|