|
chr11_+_12652427
|
2.949
|
NM_021961
|
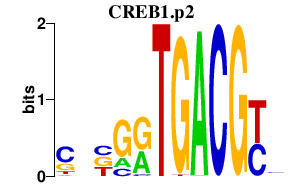
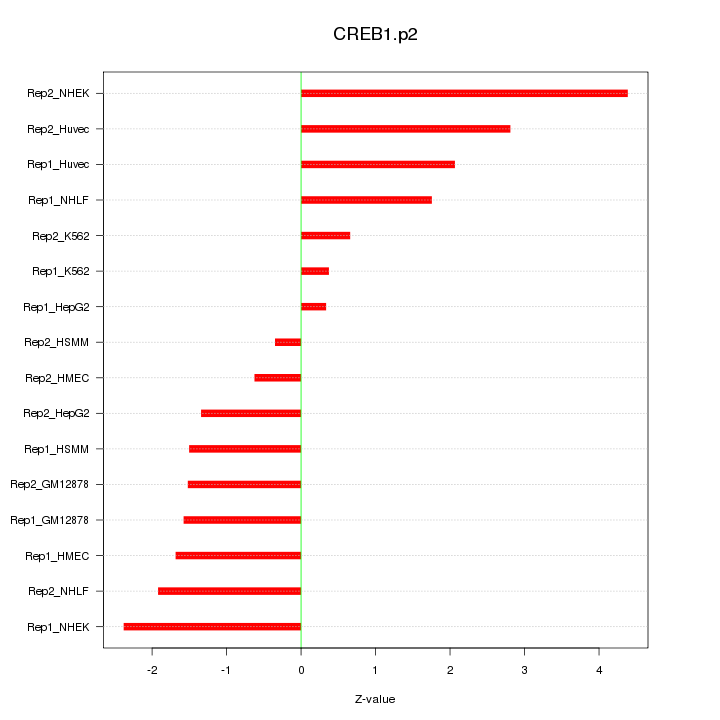
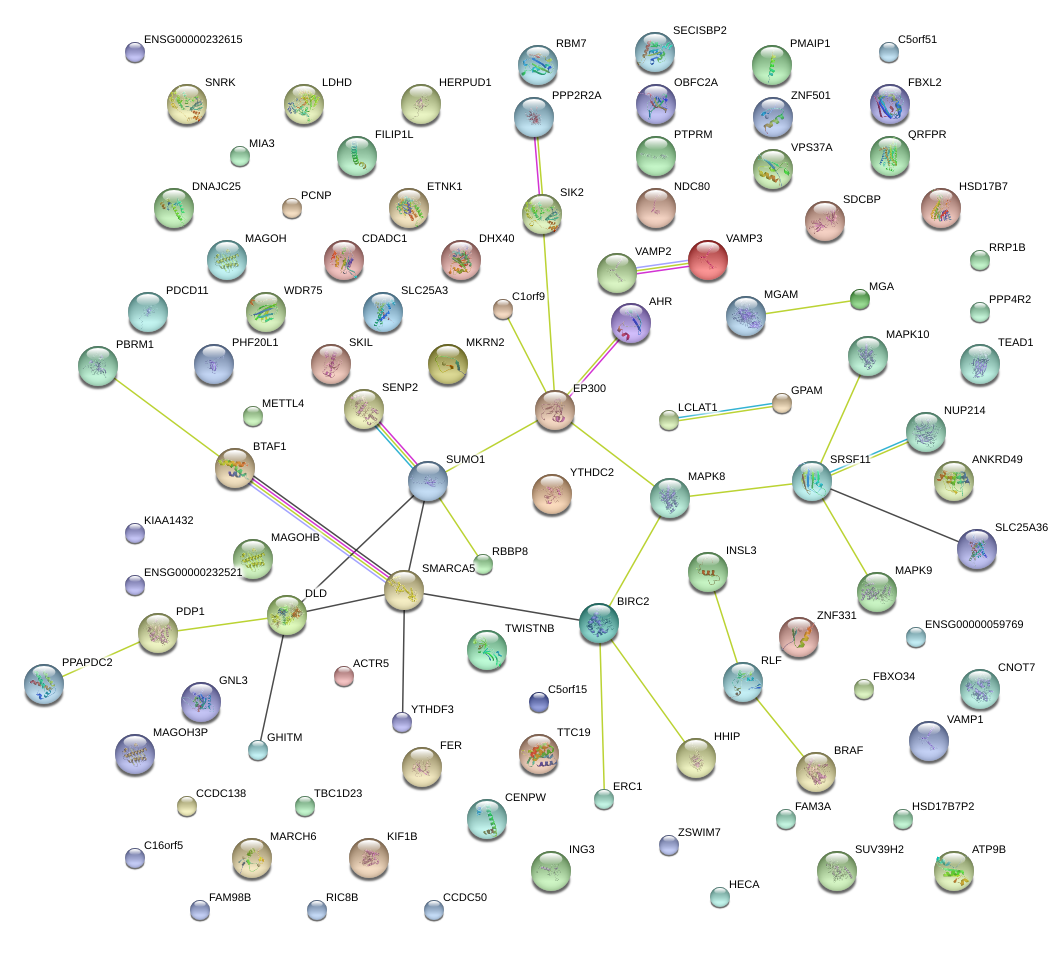
TEAD1
|
TEA domain family member 1 (SV40 transcriptional enhancer factor)
|
|
chr3_+_171558188
|
2.455
|
|
SKIL
|
SKI-like oncogene
|
|
chr3_+_171558215
|
2.404
|
|
SKIL
|
SKI-like oncogene
|
|
chr3_+_171558143
|
2.120
|
NM_001145098
NM_005414
|
SKIL
|
SKI-like oncogene
|
|
chr10_+_85889164
|
2.082
|
NM_014394
|
GHITM
|
growth hormone inducible transmembrane protein
|
|
chr10_+_49184658
|
1.986
|
|
MAPK8
|
mitogen-activated protein kinase 8
|
|
chr17_+_15843832
|
1.797
|
|
TTC19
|
tetratricopeptide repeat domain 19
|
|
chr1_+_220857774
|
1.721
|
NM_198551
|
MIA3
|
melanoma inhibitory activity family, member 3
|
|
chr3_+_43303005
|
1.683
|
NM_001100594
NM_017719
|
SNRK
|
SNF related kinase
|
|
chr2_+_192251105
|
1.636
|
NM_001031716
|
OBFC2A
|
oligonucleotide/oligosaccharide-binding fold containing 2A
|
|
chr5_+_41940216
|
1.595
|
NM_175921
|
C5orf51
|
chromosome 5 open reading frame 51
|
|
chr7_+_120378052
|
1.570
|
NM_019071
NM_198267
|
ING3
|
inhibitor of growth family, member 3
|
|
chr4_+_144654005
|
1.523
|
NM_003601
|
SMARCA5
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 5
|
|
chr8_-_17148637
|
1.518
|
NM_013354
NM_054026
|
CNOT7
|
CCR4-NOT transcription complex, subunit 7
|
|
chr12_+_970613
|
1.510
|
NM_178039
NM_178040
|
ERC1
|
ELKS/RAB6-interacting/CAST family member 1
|
|
chr4_-_120595218
|
1.452
|
|
|
|
|
chr9_+_113433364
|
1.367
|
NM_001015882
NM_004125
|
DNAJC25
DNAJC25-GNG10
|
DnaJ (Hsp40) homolog, subfamily C , member 25
DNAJC25-GNG10 readthrough
|
|
chr3_+_44746101
|
1.338
|
NM_145044
|
ZNF501
|
zinc finger protein 501
|
|
chr4_+_144654526
|
1.302
|
|
|
|
|
chr21_+_43903803
|
1.273
|
NM_015056
|
RRP1B
|
ribosomal RNA processing 1 homolog B (S. cerevisiae)
|
|
chr8_+_17148755
|
1.269
|
NM_001145152
NM_152415
|
VPS37A
|
vacuolar protein sorting 37 homolog A (S. cerevisiae)
|
|
chr7_-_19715127
|
1.265
|
NM_001002926
|
TWISTNB
|
TWIST neighbor
|
|
chr17_+_54997667
|
1.246
|
NM_001166301
NM_024612
|
DHX40
|
DEAH (Asp-Glu-Ala-His) box polypeptide 40
|
|
chr11_+_101723125
|
1.233
|
|
BIRC2
|
baculoviral IAP repeat containing 2
|
|
chr12_+_105692466
|
1.229
|
NM_018157
|
RIC8B
|
resistance to inhibitors of cholinesterase 8 homolog B (C. elegans)
|
|
chr18_+_18767275
|
1.211
|
NM_002894
|
RBBP8
|
retinoblastoma binding protein 8
|
|
chr3_+_101462375
|
1.189
|
NM_018309
|
TBC1D23
|
TBC1 domain family, member 23
|
|
chr5_+_108112537
|
1.186
|
|
FER
|
fer (fps/fes related) tyrosine kinase
|
|
chr11_+_101723157
|
1.172
|
NM_001166
|
BIRC2
|
baculoviral IAP repeat containing 2
|
|
chr8_+_59628352
|
1.165
|
|
SDCBP
|
syndecan binding protein (syntenin)
|
|
chr18_+_74930379
|
1.165
|
NM_198531
|
ATP9B
|
ATPase, class II, type 9B
|
|
chr8_+_59628372
|
1.162
|
|
SDCBP
|
syndecan binding protein (syntenin)
|
|
chr1_+_169721260
|
1.149
|
NM_015172
|
PRRC2C
|
proline-rich coiled-coil 2C
|
|
chr8_+_59628337
|
1.148
|
|
SDCBP
|
syndecan binding protein (syntenin)
|
|
chr3_+_186786655
|
1.132
|
NM_021627
|
SENP2
|
SUMO1/sentrin/SMT3 specific peptidase 2
|
|
chr18_+_2561509
|
1.131
|
NM_006101
|
NDC80
|
NDC80 homolog, kinetochore complex component (S. cerevisiae)
|
|
chr8_+_26204997
|
1.121
|
|
PPP2R2A
|
protein phosphatase 2, regulatory subunit B, alpha
|
|
chr7_-_140270749
|
1.118
|
NM_004333
|
BRAF
|
v-raf murine sarcoma viral oncogene homolog B1
|
|
chr12_+_22669352
|
1.112
|
|
ETNK1
|
ethanolamine kinase 1
|
|
chr8_+_59628281
|
1.109
|
NM_001007067
NM_001007068
NM_001007069
NM_001007070
NM_005625
|
SDCBP
|
syndecan binding protein (syntenin)
|
|
chr8_+_64243662
|
1.106
|
NM_152758
|
YTHDF3
|
YTH domain family, member 3
|
|
chr8_-_17148474
|
1.095
|
|
CNOT7
|
CCR4-NOT transcription complex, subunit 7
|
|
chr8_+_64243741
|
1.092
|
|
YTHDF3
|
YTH domain family, member 3
|
|
chr6_+_139497933
|
1.081
|
NM_016217
|
HECA
|
headcase homolog (Drosophila)
|
|
chr3_+_52694975
|
1.073
|
NM_014366
NM_206825
NM_206826
|
GNL3
|
guanine nucleotide binding protein-like 3 (nucleolar)
|
|
chr8_+_26204921
|
1.048
|
NM_002717
|
PPP2R2A
|
protein phosphatase 2, regulatory subunit B, alpha
|
|
chr18_+_2561587
|
1.030
|
|
NDC80
|
NDC80 homolog, kinetochore complex component (S. cerevisiae)
|
|
chr16_+_55523526
|
1.028
|
|
HERPUD1
|
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1
|
|
chr4_+_119419354
|
1.025
|
|
SNHG8
|
small nucleolar RNA host gene 8 (non-protein coding)
|
|
chr8_+_26204960
|
1.023
|
|
PPP2R2A
|
protein phosphatase 2, regulatory subunit B, alpha
|
|
chr12_+_97511805
|
1.011
|
|
SLC25A3
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 3
|
|
chr12_+_22669275
|
1.009
|
NM_001039481
NM_018638
|
ETNK1
|
ethanolamine kinase 1
|
|
chr19_+_58750305
|
1.005
|
NM_001079907
|
ZNF331
|
zinc finger protein 331
|
|
chr5_+_10406980
|
1.003
|
|
|
|
|
chr2_+_30523586
|
0.998
|
NM_182551
NM_001002257
|
LCLAT1
|
lysocardiolipin acyltransferase 1
|
|
chr10_+_14960787
|
0.998
|
NM_001193424
NM_001193426
NM_001193425
NM_001193427
NM_024670
|
SUV39H2
|
suppressor of variegation 3-9 homolog 2 (Drosophila)
|
|
chr18_+_74930398
|
0.995
|
|
ATP9B
|
ATPase, class II, type 9B
|
|
chr17_-_15843632
|
0.994
|
NM_001042697
NM_001042698
|
ZSWIM7
|
zinc finger, SWIM-type containing 7
|
|
chr10_+_93673544
|
0.986
|
NM_003972
|
BTAF1
|
BTAF1 RNA polymerase II, B-TFIID transcription factor-associated, 170kDa (Mot1 homolog, S. cerevisiae)
|
|
chr3_+_12573513
|
0.972
|
NM_014160
|
MKRN2
|
makorin ring finger protein 2
|
|
chr16_-_4528380
|
0.967
|
NM_001199054
|
C16orf5
|
chromosome 16 open reading frame 5
|
|
chr5_-_133332173
|
0.959
|
NM_020199
|
C5orf15
|
chromosome 5 open reading frame 15
|
|
chr14_+_54807773
|
0.958
|
NM_017943
|
FBXO34
|
F-box protein 34
|
|
chr9_+_91123204
|
0.956
|
NM_024077
|
SECISBP2
|
SECIS binding protein 2
|
|
chr2_+_108769572
|
0.955
|
NM_144978
|
CCDC138
|
coiled-coil domain containing 138
|
|
chr4_-_122521562
|
0.952
|
NM_198179
|
QRFPR
|
pyroglutamylated RFamide peptide receptor
|
|
chr8_+_94998258
|
0.951
|
NM_018444
NM_001161780
NM_001161779
NM_001161781
|
PDP1
|
pyruvate dehyrogenase phosphatase catalytic subunit 1
|
|
chr18_+_55718171
|
0.950
|
NM_021127
|
PMAIP1
|
phorbol-12-myristate-13-acetate-induced protein 1
|
|
chr11_+_113776582
|
0.936
|
NM_016090
|
RBM7
|
RNA binding motif protein 7
|
|
chr4_+_153676857
|
0.930
|
|
DKFZP434I0714
|
hypothetical protein DKFZP434I0714
|
|
chr10_+_38685299
|
0.895
|
|
HSD17B7P2
|
hydroxysteroid (17-beta) dehydrogenase 7 pseudogene 2
|
|
chr15_+_39739864
|
0.893
|
NM_001080541
NM_001164273
|
MGA
|
MAX gene associated
|
|
chr1_+_7753913
|
0.890
|
NM_004781
|
VAMP3
|
vesicle-associated membrane protein 3 (cellubrevin)
|
|
chr1_+_40399618
|
0.887
|
NM_012421
|
RLF
|
rearranged L-myc fusion
|
|
chr10_+_93673485
|
0.883
|
|
BTAF1
|
BTAF1 RNA polymerase II, B-TFIID transcription factor-associated, 170kDa (Mot1 homolog, S. cerevisiae)
|
|
chr18_+_7557313
|
0.881
|
NM_001105244
NM_002845
|
PTPRM
|
protein tyrosine phosphatase, receptor type, M
|
|
chr15_+_36533595
|
0.876
|
NM_001042429
NM_173611
|
FAM98B
|
family with sequence similarity 98, member B
|
|
chr1_+_170768635
|
0.866
|
NM_014283
|
C1orf9
|
chromosome 1 open reading frame 9
|
|
chr1_+_10193267
|
0.866
|
NM_015074
NM_183416
|
KIF1B
|
kinesin family member 1B
|
|
chr14_+_54807855
|
0.864
|
|
FBXO34
|
F-box protein 34
|
|
chr5_+_112877251
|
0.863
|
NM_022828
|
YTHDC2
|
YTH domain containing 2
|
|
chr3_+_142143329
|
0.861
|
NM_001104647
NM_018155
|
SLC25A36
|
solute carrier family 25, member 36
|
|
chr8_+_133856793
|
0.854
|
|
PHF20L1
|
PHD finger protein 20-like 1
|
|
chr3_-_52694554
|
0.852
|
NM_018313
|
PBRM1
|
polybromo 1
|
|
chr7_+_107318821
|
0.846
|
NM_000108
|
DLD
|
dihydrolipoamide dehydrogenase
|
|
chr1_+_40399642
|
0.845
|
|
RLF
|
rearranged L-myc fusion
|
|
chr3_+_184648089
|
0.842
|
|
LOC100505687
|
hypothetical LOC100505687
|
|
chr2_+_190014403
|
0.840
|
NM_032168
|
WDR75
|
WD repeat domain 75
|
|
chr3_-_101077605
|
0.839
|
NM_014890
|
FILIP1L
|
filamin A interacting protein 1-like
|
|
chr12_-_10657397
|
0.836
|
NM_018048
|
MAGOHB
|
mago-nashi homolog B (Drosophila)
|
|
chr7_+_17304707
|
0.830
|
NM_001621
|
AHR
|
aryl hydrocarbon receptor
|
|
chr17_+_15843418
|
0.826
|
NM_017775
|
TTC19
|
tetratricopeptide repeat domain 19
|
|
chr9_+_5619326
|
0.823
|
NM_001135920
|
KIAA1432
|
KIAA1432
|
|
chr12_+_97511585
|
0.821
|
|
SLC25A3
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 3
|
|
chr22_+_39817733
|
0.820
|
|
EP300
|
E1A binding protein p300
|
|
chr11_+_110978309
|
0.819
|
NM_015191
|
SIK2
|
salt-inducible kinase 2
|
|
chr9_+_132990834
|
0.814
|
|
NUP214
|
nucleoporin 214kDa
|
|
chr3_+_102775716
|
0.814
|
NM_020357
|
PCNP
|
PEST proteolytic signal containing nuclear protein
|
|
chr3_+_192529567
|
0.812
|
NM_174908
NM_178335
|
CCDC50
|
coiled-coil domain containing 50
|
|
chr4_+_145786622
|
0.811
|
NM_022475
|
HHIP
|
hedgehog interacting protein
|
|
chr3_+_33293915
|
0.806
|
NM_012157
NM_001171713
|
FBXL2
|
F-box and leucine-rich repeat protein 2
|
|
chr10_-_113933457
|
0.804
|
NM_020918
|
GPAM
|
glycerol-3-phosphate acyltransferase, mitochondrial
|
|
chr20_+_36810510
|
0.803
|
NM_024855
|
ACTR5
|
ARP5 actin-related protein 5 homolog (yeast)
|
|
chr12_+_97511596
|
0.802
|
|
SLC25A3
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 3
|
|
chr5_+_10406829
|
0.793
|
|
MARCH6
|
membrane-associated ring finger (C3HC4) 6
|
|
chr1_+_70459592
|
0.792
|
|
SRSF11
|
serine/arginine-rich splicing factor 11
|
|
chr18_-_2561422
|
0.787
|
NM_022840
|
METTL4
|
methyltransferase like 4
|
|
chr13_+_48720047
|
0.782
|
NM_001193478
NM_030911
|
CDADC1
|
cytidine and dCMP deaminase domain containing 1
|
|
chr11_+_93866790
|
0.781
|
NM_017704
|
ANKRD49
|
ankyrin repeat domain 49
|
|
chr7_+_107318852
|
0.780
|
|
DLD
|
dihydrolipoamide dehydrogenase
|
|
chr12_+_97511594
|
0.776
|
|
SLC25A3
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 3
|
|
chr6_+_126702914
|
0.774
|
NM_001012507
|
CENPW
|
centromere protein W
|
|
chr3_+_73128629
|
0.773
|
|
PPP4R2
|
protein phosphatase 4, regulatory subunit 2
|
|
chr15_+_39739933
|
0.773
|
|
MGA
|
MAX gene associated
|
|
chr7_+_107318854
|
0.772
|
|
DLD
|
dihydrolipoamide dehydrogenase
|
|
chr1_+_160797853
|
0.770
|
NM_003115
|
UAP1
|
UDP-N-acteylglucosamine pyrophosphorylase 1
|
|
chr4_-_123010918
|
0.770
|
NM_018190
NM_176824
|
BBS7
|
Bardet-Biedl syndrome 7
|
|
chr12_+_97511631
|
0.769
|
|
SLC25A3
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 3
|
|
chr2_+_187059277
|
0.769
|
|
ZC3H15
|
zinc finger CCCH-type containing 15
|
|
chr4_-_53926764
|
0.768
|
NM_152540
|
SCFD2
|
sec1 family domain containing 2
|
|
chr2_+_227408914
|
0.768
|
NM_001167608
NM_032276
|
RHBDD1
|
rhomboid domain containing 1
|
|
chr20_+_39090875
|
0.762
|
NM_003286
|
TOP1
|
topoisomerase (DNA) I
|
|
chr12_+_97511569
|
0.761
|
|
SLC25A3
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 3
|
|
chr1_+_170768914
|
0.758
|
NM_016227
|
C1orf9
|
chromosome 1 open reading frame 9
|
|
chr9_+_129414287
|
0.753
|
NM_001032221
NM_003165
|
STXBP1
|
syntaxin binding protein 1
|
|
chr4_+_55956877
|
0.752
|
|
TMEM165
|
transmembrane protein 165
|
|
chr12_+_97511520
|
0.751
|
NM_002635
NM_005888
NM_213611
|
SLC25A3
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 3
|
|
chr14_+_59785739
|
0.749
|
|
PPM1A
|
protein phosphatase, Mg2+/Mn2+ dependent, 1A
|
|
chr14_-_54563554
|
0.748
|
NM_001008396
NM_007086
|
WDHD1
|
WD repeat and HMG-box DNA binding protein 1
|
|
chr17_-_46553083
|
0.745
|
NM_001130528
NM_003971
|
SPAG9
|
sperm associated antigen 9
|
|
chr1_+_110328955
|
0.744
|
|
AHCYL1
|
adenosylhomocysteinase-like 1
|
|
chr4_+_170778303
|
0.738
|
|
CLCN3
|
chloride channel 3
|
|
chr16_+_51722285
|
0.734
|
|
CHD9
|
chromodomain helicase DNA binding protein 9
|
|
chr16_-_4528739
|
0.731
|
NM_001199055
NM_001199056
NM_013399
|
C16orf5
|
chromosome 16 open reading frame 5
|
|
chr3_+_186483408
|
0.730
|
|
LOC100505749
MAP3K13
|
hypothetical LOC100505749
mitogen-activated protein kinase kinase kinase 13
|
|
chr8_+_133856738
|
0.730
|
NM_016018
NM_032205
NM_198513
|
PHF20L1
|
PHD finger protein 20-like 1
|
|
chr19_+_44524972
|
0.727
|
|
SAMD4B
|
sterile alpha motif domain containing 4B
|
|
chr5_+_44844724
|
0.720
|
NM_016640
|
MRPS30
|
mitochondrial ribosomal protein S30
|
|
chr17_+_25828546
|
0.717
|
NM_001007024
NM_001007025
NM_004871
|
GOSR1
|
golgi SNAP receptor complex member 1
|
|
chr17_+_8279860
|
0.716
|
NM_001025579
NM_030808
|
NDEL1
|
nudE nuclear distribution gene E homolog (A. nidulans)-like 1
|
|
chr9_+_132990760
|
0.715
|
NM_005085
|
NUP214
|
nucleoporin 214kDa
|
|
chr9_+_35528890
|
0.715
|
|
RUSC2
|
RUN and SH3 domain containing 2
|
|
chr5_+_43157398
|
0.714
|
NM_003432
|
ZNF131
|
zinc finger protein 131
|
|
chr3_+_73128667
|
0.711
|
|
PPP4R2
|
protein phosphatase 4, regulatory subunit 2
|
|
chr14_-_54438913
|
0.711
|
NM_000161
NM_001024024
NM_001024070
NM_001024071
|
GCH1
|
GTP cyclohydrolase 1
|
|
chr2_+_187059237
|
0.708
|
|
ZC3H15
|
zinc finger CCCH-type containing 15
|
|
chr11_+_13646806
|
0.707
|
|
FAR1
|
fatty acyl CoA reductase 1
|
|
chr11_-_93866554
|
0.705
|
NM_005590
NM_005591
|
MRE11A
|
MRE11 meiotic recombination 11 homolog A (S. cerevisiae)
|
|
chr1_+_100276238
|
0.704
|
|
HIAT1
|
hippocampus abundant transcript 1
|
|
chr5_-_54639158
|
0.704
|
NM_019030
|
DHX29
|
DEAH (Asp-Glu-Ala-His) box polypeptide 29
|
|
chr10_+_23768203
|
0.702
|
NM_001145373
|
OTUD1
|
OTU domain containing 1
|
|
chr19_+_44524939
|
0.702
|
NM_018028
|
SAMD4B
|
sterile alpha motif domain containing 4B
|
|
chr1_+_160797973
|
0.701
|
|
UAP1
|
UDP-N-acteylglucosamine pyrophosphorylase 1
|
|
chr1_+_178867796
|
0.701
|
|
XPR1
|
xenotropic and polytropic retrovirus receptor 1
|
|
chr4_+_170778265
|
0.696
|
NM_001829
NM_173872
|
CLCN3
|
chloride channel 3
|
|
chr10_-_38305458
|
0.693
|
NM_145011
|
ZNF25
|
zinc finger protein 25
|
|
chr1_+_110328941
|
0.692
|
|
AHCYL1
|
adenosylhomocysteinase-like 1
|
|
chr3_+_73128771
|
0.692
|
NM_174907
|
PPP4R2
|
protein phosphatase 4, regulatory subunit 2
|
|
chr11_+_113776504
|
0.691
|
|
RBM7
|
RNA binding motif protein 7
|
|
chr11_-_95762632
|
0.691
|
NM_024725
|
CCDC82
|
coiled-coil domain containing 82
|
|
chr5_-_10406690
|
0.684
|
|
|
|
|
chr14_+_59785723
|
0.684
|
|
PPM1A
|
protein phosphatase, Mg2+/Mn2+ dependent, 1A
|
|
chr11_+_69794487
|
0.683
|
|
PPFIA1
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 1
|
|
chr14_+_59785703
|
0.682
|
NM_021003
NM_177951
|
PPM1A
|
protein phosphatase, Mg2+/Mn2+ dependent, 1A
|
|
chr3_+_184898234
|
0.682
|
NM_018023
|
YEATS2
|
YEATS domain containing 2
|
|
chr9_+_70046658
|
0.680
|
NM_201453
|
CBWD5
CBWD3
CBWD1
CBWD6
|
COBW domain containing 5
COBW domain containing 3
COBW domain containing 1
COBW domain containing 6
|
|
chr19_-_9407154
|
0.673
|
NM_006631
NM_198058
|
ZNF266
|
zinc finger protein 266
|
|
chr3_+_102775677
|
0.671
|
|
PCNP
|
PEST proteolytic signal containing nuclear protein
|
|
chr17_+_2443652
|
0.671
|
NM_000430
|
PAFAH1B1
|
platelet-activating factor acetylhydrolase 1b, regulatory subunit 1 (45kDa)
|
|
chr3_+_16281670
|
0.670
|
NM_138381
|
OXNAD1
|
oxidoreductase NAD-binding domain containing 1
|
|
chr6_-_38715901
|
0.665
|
NM_001099272
NM_052893
|
BTBD9
|
BTB (POZ) domain containing 9
|
|
chr5_+_95008236
|
0.664
|
NM_001131066
NM_001131065
NM_173362
|
RFESD
|
Rieske (Fe-S) domain containing
|
|
chr1_+_204875671
|
0.663
|
|
DYRK3
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 3
|
|
chr12_+_10256682
|
0.659
|
NM_031412
|
GABARAPL1
|
GABA(A) receptor-associated protein like 1
|
|
chr1_+_204875374
|
0.655
|
NM_001004023
NM_003582
|
DYRK3
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 3
|
|
chr14_+_69148062
|
0.653
|
NM_014734
|
KIAA0247
|
KIAA0247
|
|
chr7_-_106991572
|
0.653
|
|
COG5
|
component of oligomeric golgi complex 5
|
|
chr14_+_101675888
|
0.653
|
NM_144574
NM_181291
NM_181302
NM_181308
|
WDR20
|
WD repeat domain 20
|
|
chr11_+_13646822
|
0.652
|
|
FAR1
|
fatty acyl CoA reductase 1
|
|
chr4_-_148824585
|
0.648
|
NM_138364
|
PRMT10
|
protein arginine methyltransferase 10 (putative)
|
|
chr14_+_61298815
|
0.647
|
NM_003082
|
SNAPC1
|
small nuclear RNA activating complex, polypeptide 1, 43kDa
|
|
chr5_+_54639552
|
0.647
|
|
SKIV2L2
|
superkiller viralicidic activity 2-like 2 (S. cerevisiae)
|
|
chr7_+_106991638
|
0.647
|
NM_181581
|
DUS4L
|
dihydrouridine synthase 4-like (S. cerevisiae)
|
|
chr15_+_56850681
|
0.642
|
NM_001040450
NM_001040453
|
FAM63B
|
family with sequence similarity 63, member B
|
|
chr4_+_146238936
|
0.639
|
|
ABCE1
|
ATP-binding cassette, sub-family E (OABP), member 1
|
|
chr20_+_57948803
|
0.638
|
NM_001190827
NM_022106
|
C20orf177
|
chromosome 20 open reading frame 177
|
|
chr5_-_54504590
|
0.637
|
NM_001145734
NM_001170402
NM_152623
|
CDC20B
|
cell division cycle 20 homolog B (S. cerevisiae)
|
|
chr11_+_120399966
|
0.634
|
NM_001130047
NM_152715
|
TBCEL
|
tubulin folding cofactor E-like
|
|
chr11_+_13646839
|
0.631
|
|
FAR1
|
fatty acyl CoA reductase 1
|
|
chr12_-_56111054
|
0.631
|
|
R3HDM2
|
R3H domain containing 2
|
|
chr4_+_85723080
|
0.630
|
NM_001263
|
CDS1
|
CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 1
|
|
chr2_+_190014432
|
0.629
|
|
WDR75
|
WD repeat domain 75
|
|
chr4_+_110574420
|
0.628
|
|
SEC24B
|
SEC24 family, member B (S. cerevisiae)
|
|
chr12_+_97511601
|
0.627
|
|
SLC25A3
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 3
|
|
chr3_+_142569988
|
0.622
|
|
ZBTB38
|
zinc finger and BTB domain containing 38
|
|
chr5_+_54639585
|
0.622
|
|
SKIV2L2
|
superkiller viralicidic activity 2-like 2 (S. cerevisiae)
|
|
chr17_+_25730048
|
0.620
|
|
CPD
|
carboxypeptidase D
|
|
chr1_+_110328850
|
0.620
|
|
AHCYL1
|
adenosylhomocysteinase-like 1
|
|
chr17_+_25730086
|
0.618
|
|
CPD
|
carboxypeptidase D
|
|
chr2_+_172252473
|
0.615
|
|
DYNC1I2
|
dynein, cytoplasmic 1, intermediate chain 2
|