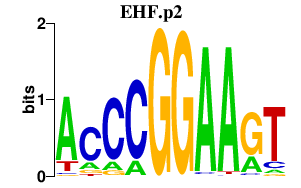
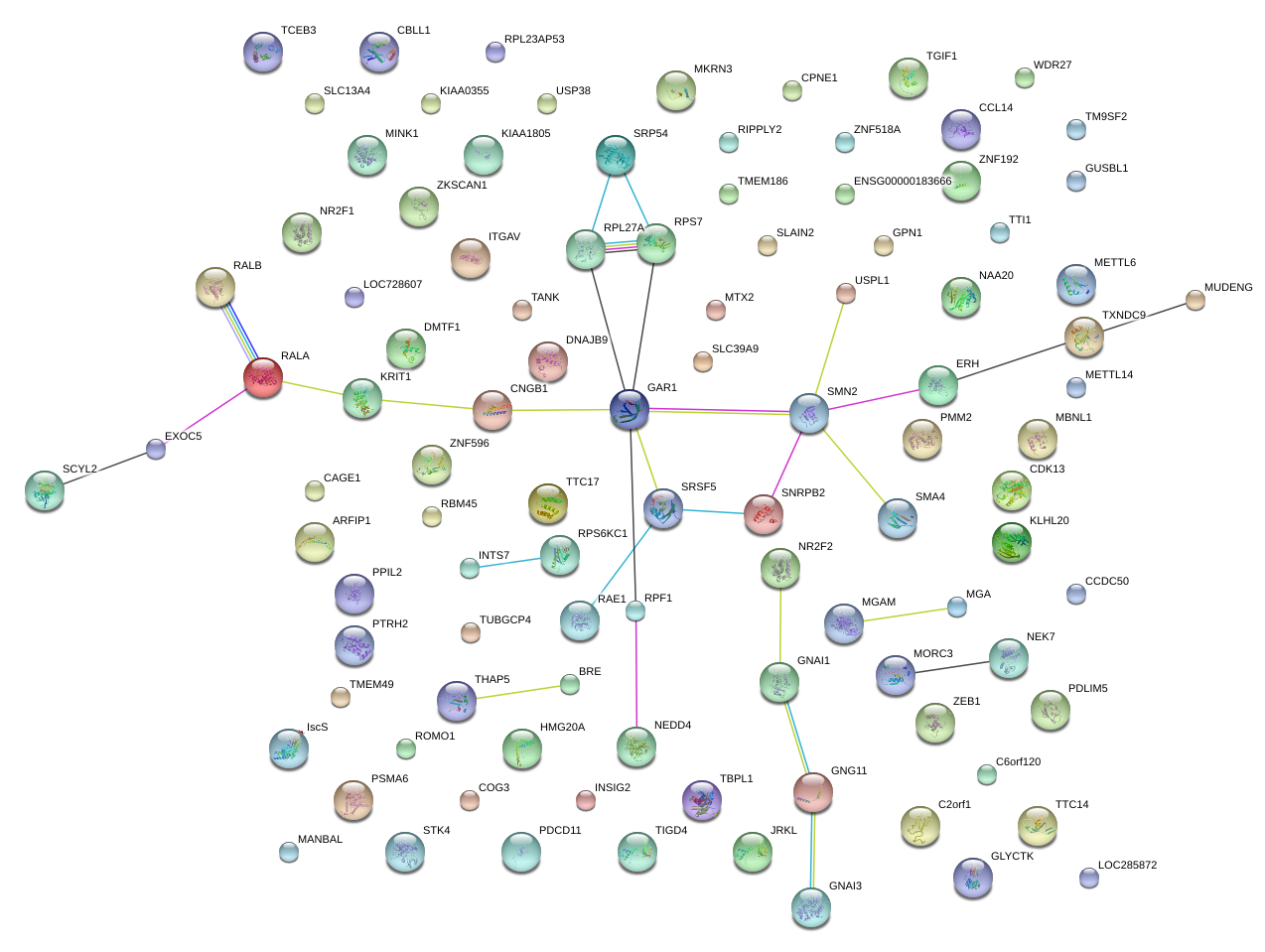
|
chr2_+_176842368
|
3.952
|
NM_006554
|
MTX2
|
metaxin 2
|
|
chr7_+_99451110
|
3.896
|
NM_003439
|
ZKSCAN1
|
zinc finger with KRAB and SCAN domains 1
|
|
chr5_+_69732561
|
3.816
|
|
SMA4
|
glucuronidase, beta pseudogene
|
|
chr7_+_107171809
|
3.672
|
|
CBLL1
|
Cas-Br-M (murine) ecotropic retroviral transforming sequence-like 1
|
|
chr2_+_118562499
|
3.655
|
NM_016133
|
INSIG2
|
insulin induced gene 2
|
|
chr6_+_134315940
|
3.631
|
NM_004865
|
TBPL1
|
TBP-like 1
|
|
chr7_+_107997527
|
3.593
|
|
DNAJB9
|
DnaJ (Hsp40) homolog, subfamily B, member 9
|
|
chr20_+_33750645
|
3.430
|
NM_080748
|
ROMO1
|
reactive oxygen species modulator 1
|
|
chr20_-_33750457
|
3.353
|
NM_001198989
NM_021100
|
NFS1
|
NFS1 nitrogen fixation 1 homolog (S. cerevisiae)
|
|
chr2_+_27659396
|
3.291
|
NM_032434
|
ZNF512
|
zinc finger protein 512
|
|
chr1_+_84717507
|
3.277
|
NM_025065
|
RPF1
|
ribosome production factor 1 homolog (S. cerevisiae)
|
|
chr20_+_16658608
|
3.226
|
NM_003092
NM_198220
|
SNRPB2
|
small nuclear ribonucleoprotein polypeptide B
|
|
chr2_+_187162969
|
3.218
|
NM_001145000
NM_002210
|
ITGAV
|
integrin, alpha V (vitronectin receptor, alpha polypeptide, antigen CD51)
|
|
chr7_-_107997132
|
3.192
|
NM_182529
NM_001130475
|
THAP5
|
THAP domain containing 5
|
|
chr4_+_144325533
|
3.140
|
|
USP38
|
ubiquitin specific peptidase 38
|
|
chr2_+_138975946
|
2.963
|
|
|
|
|
chr1_+_196392513
|
2.921
|
NM_133494
|
NEK7
|
NIMA (never in mitosis gene a)-related kinase 7
|
|
chr4_+_144325502
|
2.909
|
NM_032557
|
USP38
|
ubiquitin specific peptidase 38
|
|
chr2_+_27848085
|
2.865
|
NM_004891
NM_145330
|
MRPL33
|
mitochondrial ribosomal protein L33
|
|
chr2_+_27848090
|
2.817
|
|
MRPL33
|
mitochondrial ribosomal protein L33
|
|
chr2_+_187163232
|
2.801
|
|
ITGAV
|
integrin, alpha V (vitronectin receptor, alpha polypeptide, antigen CD51)
|
|
chr17_+_55139807
|
2.734
|
|
VMP1
|
vacuole membrane protein 1
|
|
chr14_+_34521945
|
2.677
|
|
SRP54
|
signal recognition particle 54kDa
|
|
chr17_+_55139792
|
2.669
|
|
VMP1
|
vacuole membrane protein 1
|
|
chr13_+_98951772
|
2.669
|
|
TM9SF2
|
transmembrane 9 superfamily member 2
|
|
chr14_+_34831294
|
2.649
|
NM_002791
|
PSMA6
|
proteasome (prosome, macropain) subunit, alpha type, 6
|
|
chr17_+_55139743
|
2.644
|
|
VMP1
|
vacuole membrane protein 1
|
|
chr17_+_55139793
|
2.609
|
|
VMP1
|
vacuole membrane protein 1
|
|
chr20_+_55359706
|
2.601
|
|
RAE1
|
RAE1 RNA export 1 homolog (S. pombe)
|
|
chr17_-_55139538
|
2.584
|
NM_016077
|
PTRH2
|
peptidyl-tRNA hydrolase 2
|
|
chr17_+_55139841
|
2.537
|
|
VMP1
|
vacuole membrane protein 1
|
|
chr14_+_34831355
|
2.492
|
|
PSMA6
|
proteasome (prosome, macropain) subunit, alpha type, 6
|
|
chr15_+_21361901
|
2.462
|
|
MKRN3
|
makorin ring finger protein 3
|
|
chr8_+_172199
|
2.333
|
NM_173539
NM_001042415
NM_001042416
|
ZNF596
|
zinc finger protein 596
|
|
chr22_+_20350272
|
2.314
|
NM_014337
NM_148175
NM_148176
|
PPIL2
|
peptidylprolyl isomerase (cyclophilin)-like 2
|
|
chr14_+_34521854
|
2.305
|
NM_001146282
NM_003136
|
SRP54
|
signal recognition particle 54kDa
|
|
chr2_-_138975682
|
2.305
|
|
|
|
|
chr15_+_39739864
|
2.295
|
NM_001080541
NM_001164273
|
MGA
|
MAX gene associated
|
|
chr13_+_30089829
|
2.280
|
NM_005800
|
USPL1
|
ubiquitin specific peptidase like 1
|
|
chr20_+_55359551
|
2.275
|
NM_003610
|
RAE1
|
RAE1 RNA export 1 homolog (S. pombe)
|
|
chr7_+_86619580
|
2.266
|
NM_001142326
NM_001142327
|
DMTF1
|
cyclin D binding myb-like transcription factor 1
|
|
chr2_+_3600721
|
2.254
|
NM_001011
|
RPS7
|
ribosomal protein S7
|
|
chr6_+_169844128
|
2.247
|
|
C6orf120
|
chromosome 6 open reading frame 120
|
|
chr4_+_119825983
|
2.235
|
NM_020961
|
METTL14
|
methyltransferase like 14
|
|
chr17_+_55139644
|
2.228
|
NM_030938
|
VMP1
|
vacuole membrane protein 1
|
|
chr15_+_94676691
|
2.224
|
NM_001145156
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr6_+_169844161
|
2.203
|
NM_001029863
|
C6orf120
|
chromosome 6 open reading frame 120
|
|
chr13_+_98951653
|
2.148
|
NM_004800
|
TM9SF2
|
transmembrane 9 superfamily member 2
|
|
chr8_-_172150
|
2.134
|
|
RPL23AP53
|
ribosomal protein L23a pseudogene 53
|
|
chr1_+_23942442
|
2.131
|
NM_003198
|
TCEB3
|
transcription elongation factor B (SIII), polypeptide 3 (110kDa, elongin A)
|
|
chr2_+_3600784
|
2.107
|
|
RPS7
|
ribosomal protein S7
|
|
chr11_+_43336979
|
2.087
|
NM_018259
|
TTC17
|
tetratricopeptide repeat domain 17
|
|
chr13_+_44937025
|
2.083
|
NM_031431
|
COG3
|
component of oligomeric golgi complex 3
|
|
chr1_+_23942548
|
2.077
|
|
TCEB3
|
transcription elongation factor B (SIII), polypeptide 3 (110kDa, elongin A)
|
|
chr18_+_3443771
|
2.075
|
NM_173211
|
TGIF1
|
TGFB-induced factor homeobox 1
|
|
chr6_+_169844186
|
2.070
|
|
C6orf120
|
chromosome 6 open reading frame 120
|
|
chr7_+_86619805
|
2.052
|
NM_021145
|
DMTF1
|
cyclin D binding myb-like transcription factor 1
|
|
chr7_+_107997544
|
2.018
|
NM_012328
|
DNAJB9
|
DnaJ (Hsp40) homolog, subfamily B, member 9
|
|
chr2_-_3600606
|
2.014
|
|
|
|
|
chr2_+_3600765
|
1.992
|
|
RPS7
|
ribosomal protein S7
|
|
chr12_+_130761585
|
1.980
|
NM_004592
|
SFSWAP
|
splicing factor, suppressor of white-apricot homolog (Drosophila)
|
|
chr11_+_8660913
|
1.974
|
|
RPL27A
|
ribosomal protein L27a
|
|
chr15_+_39739933
|
1.962
|
|
MGA
|
MAX gene associated
|
|
chr6_+_169844213
|
1.948
|
|
C6orf120
|
chromosome 6 open reading frame 120
|
|
chr7_-_91713232
|
1.918
|
NM_194455
NM_194456
|
KRIT1
|
KRIT1, ankyrin repeat containing
|
|
chr10_+_97879645
|
1.875
|
|
ZNF518A
|
zinc finger protein 518A
|
|
chr14_+_68934855
|
1.869
|
|
SLC39A9
|
solute carrier family 39 (zinc transporter), member 9
|
|
chr16_+_8799170
|
1.859
|
NM_000303
|
PMM2
|
phosphomannomutase 2
|
|
chr15_+_41450568
|
1.830
|
NM_014444
|
TUBGCP4
|
tubulin, gamma complex associated protein 4
|
|
chr1_+_211291153
|
1.799
|
NM_001136138
NM_012424
|
RPS6KC1
|
ribosomal protein S6 kinase, 52kDa, polypeptide 1
|
|
chr4_+_153920382
|
1.799
|
NM_001025593
NM_001025595
NM_014447
|
ARFIP1
|
ADP-ribosylation factor interacting protein 1
|
|
chr6_-_7334940
|
1.795
|
NM_001170692
NM_001170693
NM_205864
|
CAGE1
|
cancer antigen 1
|
|
chr4_+_153920563
|
1.793
|
|
ARFIP1
|
ADP-ribosylation factor interacting protein 1
|
|
chr4_-_153920326
|
1.785
|
NM_145720
|
TIGD4
|
tigger transposable element derived 4
|
|
chr1_+_211291274
|
1.783
|
|
RPS6KC1
|
ribosomal protein S6 kinase, 52kDa, polypeptide 1
|
|
chr1_+_211291334
|
1.780
|
|
RPS6KC1
|
ribosomal protein S6 kinase, 52kDa, polypeptide 1
|
|
chr14_-_56805367
|
1.762
|
NM_006544
|
EXOC5
|
exocyst complex component 5
|
|
chr11_+_95762768
|
1.752
|
NM_003772
|
JRKL
|
jerky homolog-like (mouse)
|
|
chr14_-_56805309
|
1.742
|
|
EXOC5
|
exocyst complex component 5
|
|
chr14_+_68935133
|
1.739
|
NM_018375
|
SLC39A9
|
solute carrier family 39 (zinc transporter), member 9
|
|
chr7_-_91713044
|
1.738
|
NM_004912
NM_001013406
NM_194454
|
KRIT1
|
KRIT1, ankyrin repeat containing
|
|
chr2_-_99319197
|
1.729
|
NM_005783
|
TXNDC9
|
thioredoxin domain containing 9
|
|
chr21_+_36614347
|
1.727
|
NM_015358
|
MORC3
|
MORC family CW-type zinc finger 3
|
|
chr4_+_48038095
|
1.712
|
|
SLAIN2
|
SLAIN motif family, member 2
|
|
chr1_+_171950684
|
1.705
|
NM_014458
|
KLHL20
|
kelch-like 20 (Drosophila)
|
|
chr6_-_169844023
|
1.690
|
NM_182552
|
WDR27
|
WD repeat domain 27
|
|
chr16_-_8798973
|
1.688
|
NM_015421
|
TMEM186
|
transmembrane protein 186
|
|
chr1_+_109892741
|
1.688
|
|
GNAI3
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 3
|
|
chr2_+_161725172
|
1.674
|
|
TANK
|
TRAF family member-associated NFKB activator
|
|
chr14_+_56805358
|
1.667
|
NM_018229
|
MUDENG
|
MU-2/AP1M2 domain containing, death-inducing
|
|
chr7_+_93388951
|
1.666
|
NM_004126
|
GNG11
|
guanine nucleotide binding protein (G protein), gamma 11
|
|
chr12_+_99185289
|
1.666
|
|
SCYL2
|
SCY1-like 2 (S. cerevisiae)
|
|
chr20_+_35351473
|
1.662
|
|
MANBAL
|
mannosidase, beta A, lysosomal-like
|
|
chr4_+_95592067
|
1.661
|
|
PDLIM5
|
PDZ and LIM domain 5
|
|
chr20_+_35351428
|
1.656
|
NM_001003897
NM_022077
|
MANBAL
|
mannosidase, beta A, lysosomal-like
|
|
chr14_+_68935172
|
1.655
|
|
SLC39A9
|
solute carrier family 39 (zinc transporter), member 9
|
|
chr14_-_68934720
|
1.653
|
NM_004450
|
ERH
|
enhancer of rudimentary homolog (Drosophila)
|
|
chr3_+_181802592
|
1.652
|
NM_001042601
NM_133462
|
TTC14
|
tetratricopeptide repeat domain 14
|
|
chr20_+_19946008
|
1.646
|
|
NAA20
|
N(alpha)-acetyltransferase 20, NatB catalytic subunit
|
|
chr20_-_36095236
|
1.635
|
NM_014657
|
TTI1
|
Tel2 interacting protein 1 homolog (S. pombe)
|
|
chr1_+_22224571
|
1.634
|
|
HSPC157
|
hypothetical LOC29092
|
|
chr1_+_109892760
|
1.615
|
|
GNAI3
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 3
|
|
chr4_+_110956085
|
1.607
|
NM_018983
NM_032993
|
GAR1
|
GAR1 ribonucleoprotein homolog (yeast)
|
|
chr10_+_97879457
|
1.591
|
NM_014803
|
ZNF518A
|
zinc finger protein 518A
|
|
chr2_+_178685396
|
1.586
|
NM_152945
|
RBM45
|
RNA binding motif protein 45
|
|
chr7_+_39956396
|
1.585
|
|
|
|
|
chr20_+_43028590
|
1.582
|
|
STK4
|
serine/threonine kinase 4
|
|
chr7_+_39956633
|
1.573
|
|
CDK13
|
cyclin-dependent kinase 13
|
|
chr12_+_99185334
|
1.564
|
NM_017988
|
SCYL2
|
SCY1-like 2 (S. cerevisiae)
|
|
chr7_+_134997751
|
1.564
|
NM_001130929
|
SLC13A4
PL-5283
|
solute carrier family 13 (sodium/sulfate symporters), member 4
PL-5283 protein
|
|
chr6_+_28217590
|
1.559
|
NM_006298
|
ZNF192
|
zinc finger protein 192
|
|
chr14_-_56805280
|
1.555
|
|
EXOC5
|
exocyst complex component 5
|
|
chr1_+_109892772
|
1.544
|
|
GNAI3
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 3
|
|
chr10_+_31650069
|
1.539
|
NM_001128128
NM_001174094
|
ZEB1
|
zinc finger E-box binding homeobox 1
|
|
chr12_+_99184977
|
1.533
|
|
SCYL2
|
SCY1-like 2 (S. cerevisiae)
|
|
chr3_+_192529567
|
1.530
|
NM_174908
NM_178335
|
CCDC50
|
coiled-coil domain containing 50
|
|
chr3_+_153469206
|
1.516
|
|
MBNL1
|
muscleblind-like (Drosophila)
|
|
chr7_+_39629665
|
1.514
|
NM_005402
|
RALA
|
v-ral simian leukemia viral oncogene homolog A (ras related)
|
|
chr4_+_95592030
|
1.510
|
NM_001011513
NM_001011515
NM_001011516
NM_006457
|
PDLIM5
|
PDZ and LIM domain 5
|
|
chr3_+_52296893
|
1.505
|
|
GLYCTK
|
glycerate kinase
|
|
chr14_+_69303563
|
1.503
|
|
SRSF5
|
serine/arginine-rich splicing factor 5
|
|
chr15_+_75500284
|
1.501
|
NM_018200
|
HMG20A
|
high-mobility group 20A
|
|
chr3_-_15443985
|
1.500
|
NM_152396
|
METTL6
|
methyltransferase like 6
|
|
chr7_+_39629724
|
1.498
|
|
RALA
|
v-ral simian leukemia viral oncogene homolog A (ras related)
|
|
chr14_+_69303556
|
1.491
|
|
SRSF5
|
serine/arginine-rich splicing factor 5
|
|
chr1_+_171950759
|
1.479
|
|
KLHL20
|
kelch-like 20 (Drosophila)
|
|
chr19_+_39437292
|
1.478
|
NM_014686
|
KIAA0355
|
KIAA0355
|
|
chr3_+_180348818
|
1.474
|
NM_006218
|
PIK3CA
|
phosphoinositide-3-kinase, catalytic, alpha polypeptide
|
|
chr7_+_39629782
|
1.474
|
|
RALA
|
v-ral simian leukemia viral oncogene homolog A (ras related)
|
|
chr1_+_109892708
|
1.474
|
NM_006496
|
GNAI3
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 3
|
|
chr2_+_242679507
|
1.463
|
|
LOC728323
|
hypothetical LOC728323
|
|
chr12_+_67366989
|
1.462
|
NM_020401
|
NUP107
|
nucleoporin 107kDa
|
|
chr6_+_28200365
|
1.460
|
NM_025231
|
ZSCAN16
|
zinc finger and SCAN domain containing 16
|
|
chr6_+_7335011
|
1.452
|
NM_031480
|
RIOK1
|
RIO kinase 1 (yeast)
|
|
chr2_+_69974581
|
1.452
|
NM_006857
|
SNRNP27
|
small nuclear ribonucleoprotein 27kDa (U4/U6.U5)
|
|
chr2_+_122124034
|
1.450
|
|
LOC254128
|
hypothetical LOC254128
|
|
chr20_+_48560326
|
1.449
|
|
PTPN1
|
protein tyrosine phosphatase, non-receptor type 1
|
|
chr7_+_91713347
|
1.447
|
NM_019004
|
ANKIB1
|
ankyrin repeat and IBR domain containing 1
|
|
chr7_+_39956128
|
1.439
|
|
CDK13
|
cyclin-dependent kinase 13
|
|
chr20_+_55359992
|
1.437
|
NM_001015885
|
RAE1
|
RAE1 RNA export 1 homolog (S. pombe)
|
|
chr1_+_210525885
|
1.433
|
|
PPP2R5A
|
protein phosphatase 2, regulatory subunit B', alpha
|
|
chr14_+_30161267
|
1.430
|
NM_016106
NM_182835
|
SCFD1
|
sec1 family domain containing 1
|
|
chr6_+_3966499
|
1.419
|
NM_003913
|
PRPF4B
|
PRP4 pre-mRNA processing factor 4 homolog B (yeast)
|
|
chr20_+_19945993
|
1.409
|
|
NAA20
|
N(alpha)-acetyltransferase 20, NatB catalytic subunit
|
|
chr6_+_147567200
|
1.406
|
NM_001127715
NM_139244
|
STXBP5
|
syntaxin binding protein 5 (tomosyn)
|
|
chr14_-_56805118
|
1.395
|
|
|
|
|
chr14_+_69303565
|
1.393
|
NM_001039465
NM_006925
|
SRSF5
|
serine/arginine-rich splicing factor 5
|
|
chr1_+_202097346
|
1.391
|
|
SNRPE
|
small nuclear ribonucleoprotein polypeptide E
|
|
chr6_-_11490487
|
1.377
|
NM_001142393
|
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9
|
|
chr5_-_159778621
|
1.377
|
NM_006425
|
SLU7
|
SLU7 splicing factor homolog (S. cerevisiae)
|
|
chr7_+_91914698
|
1.376
|
NM_021167
|
GATAD1
|
GATA zinc finger domain containing 1
|
|
chr20_+_36095263
|
1.374
|
NM_021215
|
RPRD1B
|
regulation of nuclear pre-mRNA domain containing 1B
|
|
chr20_+_19945949
|
1.372
|
|
NAA20
|
N(alpha)-acetyltransferase 20, NatB catalytic subunit
|
|
chr1_+_67168470
|
1.368
|
NM_001077701
NM_001077704
|
MIER1
|
mesoderm induction early response 1 homolog (Xenopus laevis)
|
|
chr3_+_52296875
|
1.366
|
NM_001144951
NM_145262
|
GLYCTK
|
glycerate kinase
|
|
chr15_+_41450642
|
1.365
|
|
TUBGCP4
|
tubulin, gamma complex associated protein 4
|
|
chr10_+_89254202
|
1.363
|
NM_001178117
NM_004897
|
MINPP1
|
multiple inositol-polyphosphate phosphatase 1
|
|
chr2_+_138438277
|
1.361
|
NM_001024074
NM_001024075
NM_006895
|
HNMT
|
histamine N-methyltransferase
|
|
chr5_+_122875691
|
1.351
|
NM_001031812
NM_001044722
NM_001044723
NM_004384
|
CSNK1G3
|
casein kinase 1, gamma 3
|
|
chr7_+_128166664
|
1.348
|
|
CALU
|
calumenin
|
|
chr7_+_128166688
|
1.336
|
|
CALU
|
calumenin
|
|
chr14_+_49157150
|
1.330
|
NM_002408
|
MGAT2
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase
|
|
chr18_-_13716582
|
1.325
|
NM_001098801
NM_152352
|
C18orf19
|
chromosome 18 open reading frame 19
|
|
chr7_+_128166636
|
1.325
|
|
CALU
|
calumenin
|
|
chr7_+_128166581
|
1.322
|
NM_001130674
NM_001219
|
CALU
|
calumenin
|
|
chr7_+_128166631
|
1.319
|
|
CALU
|
calumenin
|
|
chr12_-_99184883
|
1.316
|
NM_152317
|
DEPDC4
|
DEP domain containing 4
|
|
chr14_+_59786047
|
1.315
|
|
PPM1A
|
protein phosphatase, Mg2+/Mn2+ dependent, 1A
|
|
chr3_+_15444056
|
1.310
|
NM_033083
|
EAF1
|
ELL associated factor 1
|
|
chr2_+_187059237
|
1.305
|
|
ZC3H15
|
zinc finger CCCH-type containing 15
|
|
chr3_-_37192720
|
1.296
|
NM_001134369
NM_006309
NM_017724
|
LRRFIP2
|
leucine rich repeat (in FLII) interacting protein 2
|
|
chr2_+_187059277
|
1.294
|
|
ZC3H15
|
zinc finger CCCH-type containing 15
|
|
chr16_+_74157777
|
1.285
|
|
GABARAPL2
|
GABA(A) receptor-associated protein-like 2
|
|
chr5_+_36912595
|
1.282
|
NM_015384
NM_133433
|
NIPBL
|
Nipped-B homolog (Drosophila)
|
|
chr2_+_187059115
|
1.273
|
NM_018471
|
ZC3H15
|
zinc finger CCCH-type containing 15
|
|
chr5_+_138705416
|
1.265
|
NM_016480
|
PAIP2
|
poly(A) binding protein interacting protein 2
|
|
chr12_+_68150332
|
1.244
|
NM_001042555
NM_006654
|
FRS2
|
fibroblast growth factor receptor substrate 2
|
|
chr17_+_42355482
|
1.242
|
NM_001012511
NM_004287
NM_054022
|
GOSR2
|
golgi SNAP receptor complex member 2
|
|
chr4_+_2783764
|
1.242
|
|
SH3BP2
|
SH3-domain binding protein 2
|
|
chr8_+_110415725
|
1.237
|
NM_001193557
NM_020189
|
ENY2
|
enhancer of yellow 2 homolog (Drosophila)
|
|
chr6_+_64403668
|
1.233
|
|
PHF3
|
PHD finger protein 3
|
|
chr7_+_39956480
|
1.233
|
NM_003718
NM_031267
|
CDK13
|
cyclin-dependent kinase 13
|
|
chr6_+_237456
|
1.231
|
|
DUSP22
|
dual specificity phosphatase 22
|
|
chr16_-_14631482
|
1.228
|
|
PARN
|
poly(A)-specific ribonuclease (deadenylation nuclease)
|
|
chr16_-_14631595
|
1.228
|
NM_001134477
NM_002582
|
PARN
|
poly(A)-specific ribonuclease (deadenylation nuclease)
|
|
chr11_+_43337042
|
1.220
|
|
TTC17
|
tetratricopeptide repeat domain 17
|
|
chr1_-_167603623
|
1.217
|
NM_013330
NM_197972
|
NME7
|
non-metastatic cells 7, protein expressed in (nucleoside-diphosphate kinase)
|
|
chr1_-_118273701
|
1.214
|
|
GDAP2
|
ganglioside induced differentiation associated protein 2
|
|
chr11_-_6459139
|
1.212
|
NM_012402
|
ARFIP2
|
ADP-ribosylation factor interacting protein 2
|
|
chr11_+_123997968
|
1.210
|
|
TBRG1
|
transforming growth factor beta regulator 1
|
|
chr20_+_19945930
|
1.208
|
NM_016100
NM_181528
|
NAA20
|
N(alpha)-acetyltransferase 20, NatB catalytic subunit
|
|
chr1_-_118273728
|
1.203
|
NM_001135589
NM_017686
|
GDAP2
|
ganglioside induced differentiation associated protein 2
|
|
chr14_+_69303614
|
1.198
|
|
SRSF5
|
serine/arginine-rich splicing factor 5
|
|
chr10_-_18988172
|
1.196
|
|
|
|
|
chr16_+_3273522
|
1.195
|
|
ZNF263
|
zinc finger protein 263
|
|
chr6_+_17708483
|
1.191
|
NM_016255
|
FAM8A1
|
family with sequence similarity 8, member A1
|
|
chr2_+_138975819
|
1.189
|
NM_001001664
|
SPOPL
|
speckle-type POZ protein-like
|
|
chr5_+_87600596
|
1.188
|
|
LOC100505894
|
hypothetical LOC100505894
|
|
chr20_+_36095552
|
1.182
|
|
RPRD1B
|
regulation of nuclear pre-mRNA domain containing 1B
|
|
chr20_+_48560327
|
1.178
|
|
PTPN1
|
protein tyrosine phosphatase, non-receptor type 1
|