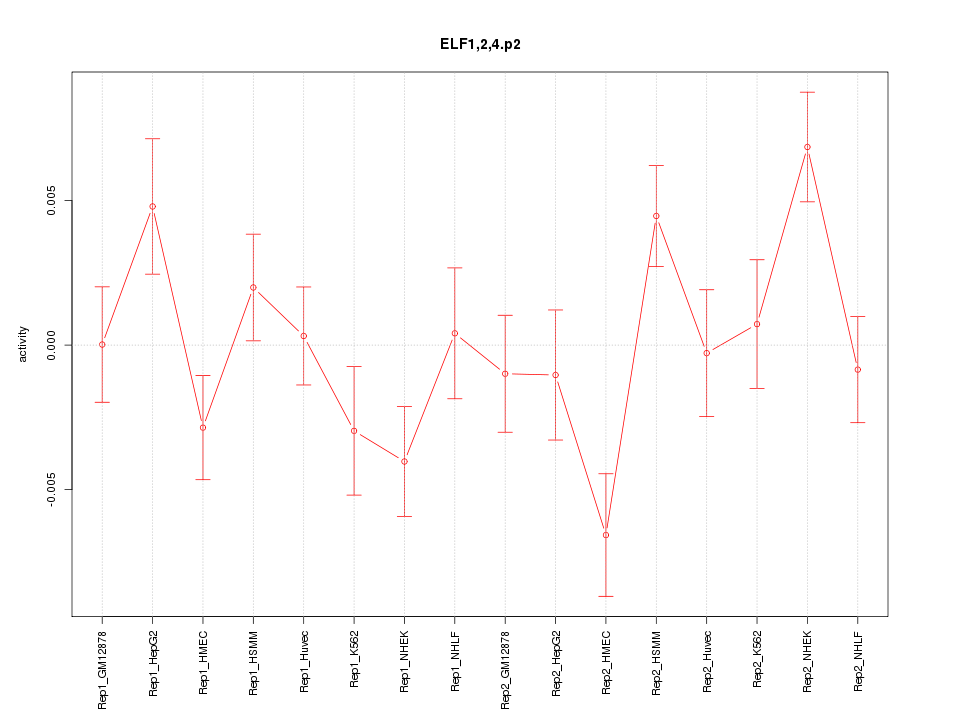
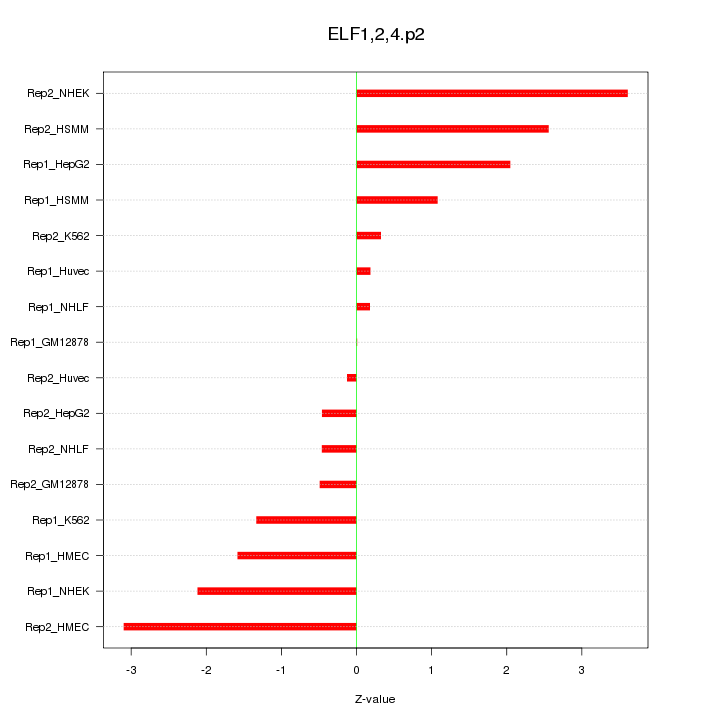
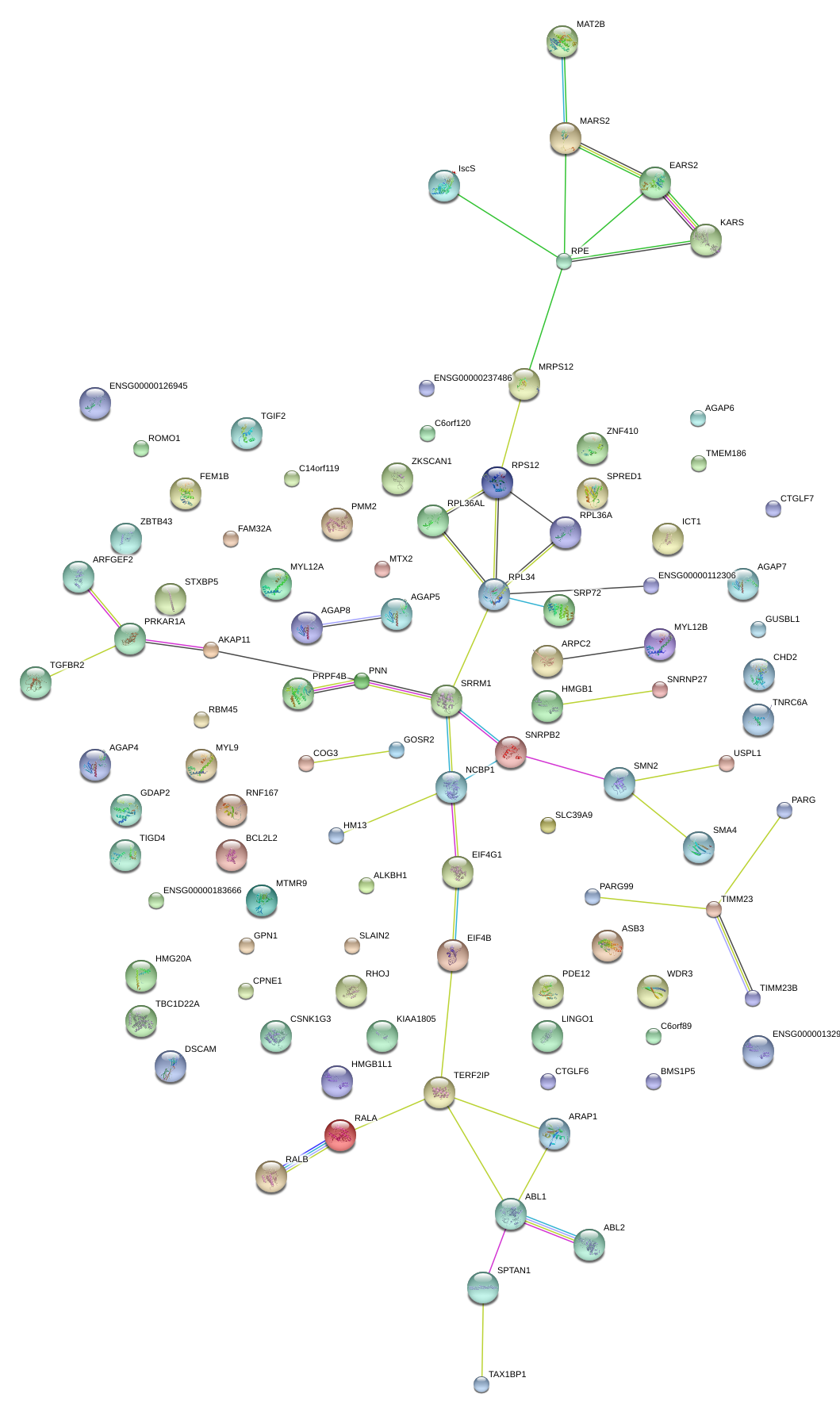
|
chr22_+_25398690
|
1.714
|
|
LOC388889
|
hypothetical LOC388889
|
|
chr17_+_42355492
|
1.525
|
|
GOSR2
|
golgi SNAP receptor complex member 2
|
|
chr5_+_122875691
|
1.373
|
NM_001031812
NM_001044722
NM_001044723
NM_004384
|
CSNK1G3
|
casein kinase 1, gamma 3
|
|
chr17_+_42355482
|
1.311
|
NM_001012511
NM_004287
NM_054022
|
GOSR2
|
golgi SNAP receptor complex member 2
|
|
chr13_-_30089509
|
1.264
|
|
HMGB1
|
high-mobility group box 1
|
|
chr13_+_30089829
|
1.201
|
NM_005800
|
USPL1
|
ubiquitin specific peptidase like 1
|
|
chr20_+_16658608
|
1.061
|
NM_003092
NM_198220
|
SNRPB2
|
small nuclear ribonucleoprotein polypeptide B
|
|
chr4_+_48038259
|
1.013
|
NM_020846
|
SLAIN2
|
SLAIN motif family, member 2
|
|
chr14_+_22845851
|
0.937
|
NM_004050
|
BCL2L2-PABPN1
BCL2L2
|
BCL2L2-PABPN1 read-through transcript
BCL2-like 2
|
|
chr6_+_36961617
|
0.935
|
NM_152734
|
C6orf89
|
chromosome 6 open reading frame 89
|
|
chr5_+_162865128
|
0.912
|
NM_013283
|
MAT2B
|
methionine adenosyltransferase II, beta
|
|
chr17_+_4784524
|
0.894
|
|
RNF167
|
ring finger protein 167
|
|
chr20_+_33750645
|
0.867
|
NM_080748
|
ROMO1
|
reactive oxygen species modulator 1
|
|
chr12_+_51686328
|
0.854
|
NM_001417
|
EIF4B
|
eukaryotic translation initiation factor 4B
|
|
chr20_-_33750457
|
0.842
|
NM_001198989
NM_021100
|
NFS1
|
NFS1 nitrogen fixation 1 homolog (S. cerevisiae)
|
|
chr10_-_51293220
|
0.834
|
NM_006327
|
TIMM23
TIMM23B
|
translocase of inner mitochondrial membrane 23 homolog (yeast)
translocase of inner mitochondrial membrane 23 homolog B (yeast)
|
|
chr15_+_91248706
|
0.821
|
|
CHD2
|
chromodomain helicase DNA binding protein 2
|
|
chr4_+_48038095
|
0.812
|
|
SLAIN2
|
SLAIN motif family, member 2
|
|
chr1_+_118273869
|
0.806
|
|
WDR3
|
WD repeat domain 3
|
|
chr20_+_29598848
|
0.795
|
|
HM13
|
histocompatibility (minor) 13
|
|
chr16_+_2593351
|
0.787
|
|
LOC652276
|
hypothetical LOC652276
|
|
chr1_-_118273701
|
0.772
|
|
GDAP2
|
ganglioside induced differentiation associated protein 2
|
|
chr2_-_53867488
|
0.768
|
NM_016115
NM_145863
|
ASB3
|
ankyrin repeat and SOCS box containing 3
|
|
chr7_+_27746335
|
0.762
|
|
TAX1BP1
|
Tax1 (human T-cell leukemia virus type I) binding protein 1
|
|
chr9_+_132579088
|
0.760
|
NM_007313
|
ABL1
|
c-abl oncogene 1, non-receptor tyrosine kinase
|
|
chr10_-_51041156
|
0.759
|
NM_003631
|
AGAP8
LOC728407
PARG
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 8
poly(ADP-ribose) glycohydrolase pseudogene
poly (ADP-ribose) glycohydrolase
|
|
chr2_+_27659396
|
0.756
|
NM_032434
|
ZNF512
|
zinc finger protein 512
|
|
chr3_+_30623376
|
0.735
|
|
TGFBR2
|
transforming growth factor, beta receptor II (70/80kDa)
|
|
chr14_+_62740878
|
0.730
|
NM_020663
|
RHOJ
|
ras homolog gene family, member J
|
|
chr7_+_27746334
|
0.729
|
|
|
|
|
chr7_+_99451110
|
0.696
|
NM_003439
|
ZKSCAN1
|
zinc finger with KRAB and SCAN domains 1
|
|
chr9_+_130354490
|
0.687
|
NM_001130438
NM_001195532
NM_003127
|
SPTAN1
|
spectrin, alpha, non-erythrocytic 1 (alpha-fodrin)
|
|
chr1_+_118273887
|
0.683
|
NM_006784
|
WDR3
|
WD repeat domain 3
|
|
chr15_-_75693309
|
0.679
|
|
LINGO1
|
leucine rich repeat and Ig domain containing 1
|
|
chr18_+_3252184
|
0.679
|
|
MYL12B
|
myosin, light chain 12B, regulatory
|
|
chr12_+_51686504
|
0.677
|
|
EIF4B
|
eukaryotic translation initiation factor 4B
|
|
chr2_+_210575600
|
0.671
|
NM_199229
|
RPE
|
ribulose-5-phosphate-3-epimerase
|
|
chr16_+_74239135
|
0.667
|
NM_018975
|
TERF2IP
|
telomeric repeat binding factor 2, interacting protein
|
|
chr2_+_210575566
|
0.662
|
NM_006916
|
RPE
|
ribulose-5-phosphate-3-epimerase
|
|
chr18_+_3252110
|
0.656
|
NM_033546
|
MYL12B
|
myosin, light chain 12B, regulatory
|
|
chr20_+_34635362
|
0.652
|
NM_021809
|
TGIF2
|
TGFB-induced factor homeobox 2
|
|
chr22_+_45537181
|
0.650
|
NM_014346
|
TBC1D22A
|
TBC1 domain family, member 22A
|
|
chr19_+_16157198
|
0.647
|
NM_014077
|
FAM32A
|
family with sequence similarity 32, member A
|
|
chr3_+_57517013
|
0.644
|
NM_177966
|
PDE12
|
phosphodiesterase 12
|
|
chr7_+_27746268
|
0.639
|
|
TAX1BP1
|
Tax1 (human T-cell leukemia virus type I) binding protein 1
|
|
chr17_+_70520339
|
0.632
|
NM_001545
|
ICT1
|
immature colon carcinoma transcript 1
|
|
chr15_+_81216349
|
0.629
|
|
|
|
|
chr14_+_68935133
|
0.628
|
NM_018375
|
SLC39A9
|
solute carrier family 39 (zinc transporter), member 9
|
|
chr15_-_81216179
|
0.627
|
|
|
|
|
chr6_+_169844186
|
0.627
|
|
C6orf120
|
chromosome 6 open reading frame 120
|
|
chr14_+_73423260
|
0.626
|
NM_021188
|
ZNF410
|
zinc finger protein 410
|
|
chr14_+_68935172
|
0.623
|
|
SLC39A9
|
solute carrier family 39 (zinc transporter), member 9
|
|
chr6_+_169844213
|
0.620
|
|
C6orf120
|
chromosome 6 open reading frame 120
|
|
chr14_+_38720085
|
0.620
|
|
PNN
|
pinin, desmosome associated protein
|
|
chr15_+_75500284
|
0.618
|
NM_018200
|
HMG20A
|
high-mobility group 20A
|
|
chr12_+_51686512
|
0.617
|
|
EIF4B
|
eukaryotic translation initiation factor 4B
|
|
chr15_+_36331411
|
0.615
|
|
SPRED1
|
sprouty-related, EVH1 domain containing 1
|
|
chr6_+_169844161
|
0.609
|
NM_001029863
|
C6orf120
|
chromosome 6 open reading frame 120
|
|
chr2_+_218790109
|
0.598
|
NM_152862
|
ARPC2
|
actin related protein 2/3 complex, subunit 2, 34kDa
|
|
chr5_+_69732561
|
0.597
|
|
SMA4
|
glucuronidase, beta pseudogene
|
|
chr4_+_109761170
|
0.592
|
NM_000995
NM_033625
|
RPL34
|
ribosomal protein L34
|
|
chr4_+_57028536
|
0.592
|
|
SRP72
|
signal recognition particle 72kDa
|
|
chr2_+_198278276
|
0.585
|
NM_138395
|
MARS2
|
methionyl-tRNA synthetase 2, mitochondrial
|
|
chr9_+_128607081
|
0.584
|
NM_001135776
NM_014007
|
ZBTB43
|
zinc finger and BTB domain containing 43
|
|
chr16_-_74239030
|
0.583
|
NM_001130089
NM_005548
|
KARS
|
lysyl-tRNA synthetase
|
|
chr3_+_57517094
|
0.582
|
|
PDE12
|
phosphodiesterase 12
|
|
chr9_+_99435900
|
0.582
|
|
NCBP1
|
nuclear cap binding protein subunit 1, 80kDa
|
|
chr16_+_8799170
|
0.581
|
NM_000303
|
PMM2
|
phosphomannomutase 2
|
|
chr7_+_39629782
|
0.581
|
|
RALA
|
v-ral simian leukemia viral oncogene homolog A (ras related)
|
|
chr6_+_133177403
|
0.575
|
|
RPS12
|
ribosomal protein S12
|
|
chr2_+_218790143
|
0.570
|
|
ARPC2
|
actin related protein 2/3 complex, subunit 2, 34kDa
|
|
chr20_-_34635091
|
0.570
|
|
|
|
|
chr13_+_44937025
|
0.570
|
NM_031431
|
COG3
|
component of oligomeric golgi complex 3
|
|
chr13_+_41744255
|
0.567
|
NM_016248
|
AKAP11
|
A kinase (PRKA) anchor protein 11
|
|
chr6_+_3966499
|
0.562
|
NM_003913
|
PRPF4B
|
PRP4 pre-mRNA processing factor 4 homolog B (yeast)
|
|
chr6_+_147567200
|
0.561
|
NM_001127715
NM_139244
|
STXBP5
|
syntaxin binding protein 5 (tomosyn)
|
|
chr15_+_66357456
|
0.549
|
|
FEM1B
|
fem-1 homolog b (C. elegans)
|
|
chr8_+_11179724
|
0.549
|
|
MTMR9
|
myotubularin related protein 9
|
|
chr17_+_64019748
|
0.545
|
|
PRKAR1A
|
protein kinase, cAMP-dependent, regulatory, type I, alpha (tissue specific extinguisher 1)
|
|
chr16_+_24648446
|
0.545
|
NM_014494
|
TNRC6A
|
trinucleotide repeat containing 6A
|
|
chr1_+_24842381
|
0.544
|
|
SRRM1
|
serine/arginine repetitive matrix 1
|
|
chr6_+_169844128
|
0.539
|
|
C6orf120
|
chromosome 6 open reading frame 120
|
|
chr16_-_8798973
|
0.538
|
NM_015421
|
TMEM186
|
transmembrane protein 186
|
|
chr14_-_77244073
|
0.538
|
NM_006020
|
ALKBH1
|
alkB, alkylation repair homolog 1 (E. coli)
|
|
chr14_-_49157076
|
0.534
|
NM_001001
|
RPL36AL
|
ribosomal protein L36a-like
|
|
chr4_-_153920326
|
0.533
|
NM_145720
|
TIGD4
|
tigger transposable element derived 4
|
|
chr2_+_176842368
|
0.530
|
NM_006554
|
MTX2
|
metaxin 2
|
|
chr14_+_22634585
|
0.529
|
|
C14orf119
|
chromosome 14 open reading frame 119
|
|
chr20_+_46971618
|
0.527
|
NM_006420
|
ARFGEF2
|
ADP-ribosylation factor guanine nucleotide-exchange factor 2 (brefeldin A-inhibited)
|
|
chr11_-_72111027
|
0.527
|
NM_001135190
NM_015242
|
ARAP1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1
|
|
chr2_+_178685396
|
0.527
|
NM_152945
|
RBM45
|
RNA binding motif protein 45
|
|
chr7_+_39629724
|
0.523
|
|
RALA
|
v-ral simian leukemia viral oncogene homolog A (ras related)
|
|
chr16_-_23476138
|
0.521
|
NM_001083614
|
EARS2
|
glutamyl-tRNA synthetase 2, mitochondrial (putative)
|
|
chr14_+_22634517
|
0.521
|
|
C14orf119
|
chromosome 14 open reading frame 119
|
|
chr2_+_69974581
|
0.517
|
NM_006857
|
SNRNP27
|
small nuclear ribonucleoprotein 27kDa (U4/U6.U5)
|
|
chr3_+_185515543
|
0.515
|
NM_182917
|
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1
|
|
chr2_-_196641682
|
0.514
|
NM_018897
|
DNAH7
|
dynein, axonemal, heavy chain 7
|
|
chr2_+_218790159
|
0.512
|
|
ARPC2
|
actin related protein 2/3 complex, subunit 2, 34kDa
|
|
chr13_+_76464044
|
0.512
|
NM_006493
|
CLN5
|
ceroid-lipofuscinosis, neuronal 5
|
|
chr15_+_65145248
|
0.511
|
NM_005902
|
SMAD3
|
SMAD family member 3
|
|
chr2_+_32435800
|
0.510
|
|
BIRC6
|
baculoviral IAP repeat containing 6
|
|
chr11_+_63363174
|
0.508
|
|
MARK2
|
MAP/microtubule affinity-regulating kinase 2
|
|
chr6_-_11490487
|
0.506
|
NM_001142393
|
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9
|
|
chr14_+_34831294
|
0.504
|
NM_002791
|
PSMA6
|
proteasome (prosome, macropain) subunit, alpha type, 6
|
|
chr14_+_34831355
|
0.504
|
|
PSMA6
|
proteasome (prosome, macropain) subunit, alpha type, 6
|
|
chr14_+_68934855
|
0.504
|
|
SLC39A9
|
solute carrier family 39 (zinc transporter), member 9
|
|
chr9_-_132444639
|
0.504
|
|
LOC100272217
|
hypothetical LOC100272217
|
|
chr2_+_65308332
|
0.503
|
NM_001005386
NM_005722
|
ACTR2
|
ARP2 actin-related protein 2 homolog (yeast)
|
|
chr3_+_133861894
|
0.503
|
|
UBA5
|
ubiquitin-like modifier activating enzyme 5
|
|
chr19_+_50150486
|
0.503
|
|
CLPTM1
|
cleft lip and palate associated transmembrane protein 1
|
|
chr14_+_77244177
|
0.501
|
NM_031210
|
C14orf156
|
chromosome 14 open reading frame 156
|
|
chr4_+_153920563
|
0.501
|
|
ARFIP1
|
ADP-ribosylation factor interacting protein 1
|
|
chr17_-_71174816
|
0.501
|
NM_004259
NM_001003715
NM_001003716
|
RECQL5
|
RecQ protein-like 5
|
|
chr6_-_169844023
|
0.497
|
NM_182552
|
WDR27
|
WD repeat domain 27
|
|
chr14_+_22634621
|
0.496
|
NM_017924
|
C14orf119
|
chromosome 14 open reading frame 119
|
|
chr1_-_118273728
|
0.496
|
NM_001135589
NM_017686
|
GDAP2
|
ganglioside induced differentiation associated protein 2
|
|
chr8_+_125620523
|
0.494
|
NM_005005
|
NDUFB9
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 9, 22kDa
|
|
chr4_+_56414565
|
0.492
|
NM_001024924
NM_018261
NM_178237
|
EXOC1
|
exocyst complex component 1
|
|
chr8_+_117847945
|
0.491
|
|
UTP23
|
UTP23, small subunit (SSU) processome component, homolog (yeast)
|
|
chr19_+_50150441
|
0.491
|
NM_001294
|
CLPTM1
|
cleft lip and palate associated transmembrane protein 1
|
|
chr1_+_109892741
|
0.490
|
|
GNAI3
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 3
|
|
chr7_+_39629665
|
0.490
|
NM_005402
|
RALA
|
v-ral simian leukemia viral oncogene homolog A (ras related)
|
|
chr14_+_96038479
|
0.483
|
|
PAPOLA
|
poly(A) polymerase alpha
|
|
chr8_+_117847922
|
0.482
|
NM_032334
|
UTP23
|
UTP23, small subunit (SSU) processome component, homolog (yeast)
|
|
chr14_-_22634662
|
0.482
|
NM_001164814
NM_001164815
NM_014977
|
ACIN1
|
apoptotic chromatin condensation inducer 1
|
|
chr17_-_24013303
|
0.481
|
|
SDF2
|
stromal cell-derived factor 2
|
|
chr16_+_79626817
|
0.481
|
NM_015251
|
ATMIN
|
ATM interactor
|
|
chr14_+_54104099
|
0.480
|
|
SAMD4A
|
sterile alpha motif domain containing 4A
|
|
chr3_+_61522219
|
0.480
|
NM_002841
|
PTPRG
|
protein tyrosine phosphatase, receptor type, G
|
|
chr12_-_61283474
|
0.477
|
NM_175895
|
C12orf61
|
chromosome 12 open reading frame 61
|
|
chr17_+_24013359
|
0.476
|
NM_003170
|
SUPT6H
|
suppressor of Ty 6 homolog (S. cerevisiae)
|
|
chr4_+_153920382
|
0.474
|
NM_001025593
NM_001025595
NM_014447
|
ARFIP1
|
ADP-ribosylation factor interacting protein 1
|
|
chr22_-_45537081
|
0.473
|
|
|
|
|
chr16_+_48865521
|
0.472
|
|
ADCY7
|
adenylate cyclase 7
|
|
chr15_+_56850776
|
0.471
|
|
FAM63B
|
family with sequence similarity 63, member B
|
|
chr11_+_74950893
|
0.471
|
|
SERPINH1
|
serpin peptidase inhibitor, clade H (heat shock protein 47), member 1, (collagen binding protein 1)
|
|
chr20_+_3138055
|
0.470
|
NM_033453
NM_181493
|
ITPA
|
inosine triphosphatase (nucleoside triphosphate pyrophosphatase)
|
|
chr12_-_99184883
|
0.470
|
NM_152317
|
DEPDC4
|
DEP domain containing 4
|
|
chr20_+_43028533
|
0.469
|
NM_006282
|
STK4
|
serine/threonine kinase 4
|
|
chr2_+_239000395
|
0.469
|
|
ASB1
|
ankyrin repeat and SOCS box containing 1
|
|
chr14_+_77244191
|
0.469
|
|
C14orf156
|
chromosome 14 open reading frame 156
|
|
chr1_+_109892760
|
0.468
|
|
GNAI3
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 3
|
|
chr1_+_65658903
|
0.466
|
NM_001003679
NM_001003680
NM_002303
NM_001198681
|
LEPR
LEPROT
|
leptin receptor
leptin receptor overlapping transcript
|
|
chr16_+_23476334
|
0.466
|
NM_019116
|
UBFD1
|
ubiquitin family domain containing 1
|
|
chr14_+_101346032
|
0.466
|
|
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma
|
|
chr2_+_53867571
|
0.465
|
NM_001127397
NM_001127398
NM_015701
|
ERLEC1
|
endoplasmic reticulum lectin 1
|
|
chr5_+_67620007
|
0.464
|
NM_181524
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha)
|
|
chr10_-_43212703
|
0.462
|
NM_001098207
|
HNRNPF
|
heterogeneous nuclear ribonucleoprotein F
|
|
chr2_+_198089036
|
0.458
|
|
MOBKL3
|
MOB1, Mps One Binder kinase activator-like 3 (yeast)
|
|
chr15_+_88610181
|
0.457
|
|
NGRN
|
neugrin, neurite outgrowth associated
|
|
chr11_-_65860450
|
0.457
|
NM_004292
|
RIN1
|
Ras and Rab interactor 1
|
|
chr11_+_63362922
|
0.456
|
NM_001039469
NM_001163296
NM_001163297
NM_004954
|
MARK2
|
MAP/microtubule affinity-regulating kinase 2
|
|
chr17_+_24013320
|
0.455
|
|
SUPT6H
|
suppressor of Ty 6 homolog (S. cerevisiae)
|
|
chr17_+_55052185
|
0.451
|
|
|
|
|
chr16_+_72888231
|
0.451
|
|
PSMD7
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 7
|
|
chr4_+_144325533
|
0.450
|
|
USP38
|
ubiquitin specific peptidase 38
|
|
chr6_+_26646548
|
0.450
|
NM_006353
|
HMGN4
|
high mobility group nucleosomal binding domain 4
|
|
chr3_+_180348818
|
0.449
|
NM_006218
|
PIK3CA
|
phosphoinositide-3-kinase, catalytic, alpha polypeptide
|
|
chr17_+_59205289
|
0.449
|
NM_007372
NM_203499
|
DDX42
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 42
|
|
chr16_+_72888161
|
0.448
|
NM_002811
|
PSMD7
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 7
|
|
chr4_+_144325502
|
0.446
|
NM_032557
|
USP38
|
ubiquitin specific peptidase 38
|
|
chr12_+_99184977
|
0.446
|
|
SCYL2
|
SCY1-like 2 (S. cerevisiae)
|
|
chr20_+_43028590
|
0.443
|
|
STK4
|
serine/threonine kinase 4
|
|
chr1_+_94656517
|
0.442
|
NM_001122674
NM_002858
|
ABCD3
|
ATP-binding cassette, sub-family D (ALD), member 3
|
|
chr9_+_4669557
|
0.442
|
NM_017913
|
CDC37L1
|
cell division cycle 37 homolog (S. cerevisiae)-like 1
|
|
chr3_+_144203061
|
0.441
|
NM_001080415
|
U2SURP
|
U2 snRNP-associated SURP domain containing
|
|
chr2_+_64605049
|
0.440
|
|
AFTPH
|
aftiphilin
|
|
chr17_+_55139644
|
0.440
|
NM_030938
|
VMP1
|
vacuole membrane protein 1
|
|
chr8_-_125620466
|
0.440
|
NM_001146160
NM_032026
|
TATDN1
|
TatD DNase domain containing 1
|
|
chr14_+_52243645
|
0.440
|
NM_002806
|
PSMC6
|
proteasome (prosome, macropain) 26S subunit, ATPase, 6
|
|
chr12_+_99185289
|
0.439
|
|
SCYL2
|
SCY1-like 2 (S. cerevisiae)
|
|
chr21_+_44256630
|
0.439
|
NM_003274
|
TRAPPC10
|
trafficking protein particle complex 10
|
|
chr2_+_118288754
|
0.439
|
|
DDX18
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 18
|
|
chr2_+_161725172
|
0.437
|
|
TANK
|
TRAF family member-associated NFKB activator
|
|
chr17_+_33761234
|
0.435
|
NM_014598
|
SOCS7
|
suppressor of cytokine signaling 7
|
|
chr12_-_52868855
|
0.434
|
NM_014311
|
SMUG1
|
single-strand-selective monofunctional uracil-DNA glycosylase 1
|
|
chr14_+_101345878
|
0.432
|
NM_002719
NM_178586
NM_178587
|
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma
|
|
chr15_+_56850681
|
0.432
|
NM_001040450
NM_001040453
|
FAM63B
|
family with sequence similarity 63, member B
|
|
chr14_-_22634134
|
0.432
|
|
ACIN1
|
apoptotic chromatin condensation inducer 1
|
|
chr3_+_140545528
|
0.430
|
NM_020191
|
MRPS22
|
mitochondrial ribosomal protein S22
|
|
chr2_+_182464713
|
0.429
|
NM_001130445
NM_006751
|
SSFA2
|
sperm specific antigen 2
|
|
chr17_-_24013259
|
0.429
|
|
SDF2
|
stromal cell-derived factor 2
|
|
chr20_-_48741481
|
0.429
|
|
|
|
|
chr5_+_61910318
|
0.428
|
NM_181506
|
LRRC70
|
leucine rich repeat containing 70
|
|
chr17_+_55139743
|
0.428
|
|
VMP1
|
vacuole membrane protein 1
|
|
chr4_+_57028503
|
0.427
|
NM_006947
|
SRP72
|
signal recognition particle 72kDa
|
|
chr3_+_61522310
|
0.427
|
|
PTPRG
|
protein tyrosine phosphatase, receptor type, G
|
|
chr17_+_59205244
|
0.426
|
|
|
|
|
chr2_-_201461945
|
0.426
|
NM_032472
NM_130906
|
PPIL3
|
peptidylprolyl isomerase (cyclophilin)-like 3
|
|
chr17_+_55139792
|
0.424
|
|
VMP1
|
vacuole membrane protein 1
|
|
chr5_+_72830029
|
0.423
|
|
BTF3
|
basic transcription factor 3
|
|
chr15_+_88609837
|
0.423
|
NM_001033088
|
NGRN
|
neugrin, neurite outgrowth associated
|
|
chr10_+_18988282
|
0.423
|
NM_178815
|
ARL5B
|
ADP-ribosylation factor-like 5B
|
|
chr1_+_65658718
|
0.422
|
NM_001198683
NM_017526
|
LEPROT
|
leptin receptor overlapping transcript
|
|
chr10_-_43212681
|
0.419
|
|
HNRNPF
|
heterogeneous nuclear ribonucleoprotein F
|
|
chr5_+_72830003
|
0.417
|
NM_001037637
NM_001207
|
BTF3
|
basic transcription factor 3
|
|
chr4_+_119825983
|
0.417
|
NM_020961
|
METTL14
|
methyltransferase like 14
|
|
chr1_+_94656556
|
0.415
|
|
ABCD3
|
ATP-binding cassette, sub-family D (ALD), member 3
|
|
chr17_+_64019693
|
0.414
|
NM_212471
|
PRKAR1A
|
protein kinase, cAMP-dependent, regulatory, type I, alpha (tissue specific extinguisher 1)
|
|
chr17_+_55139793
|
0.413
|
|
VMP1
|
vacuole membrane protein 1
|