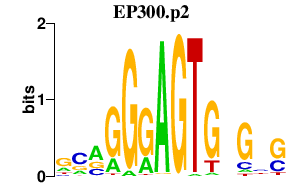
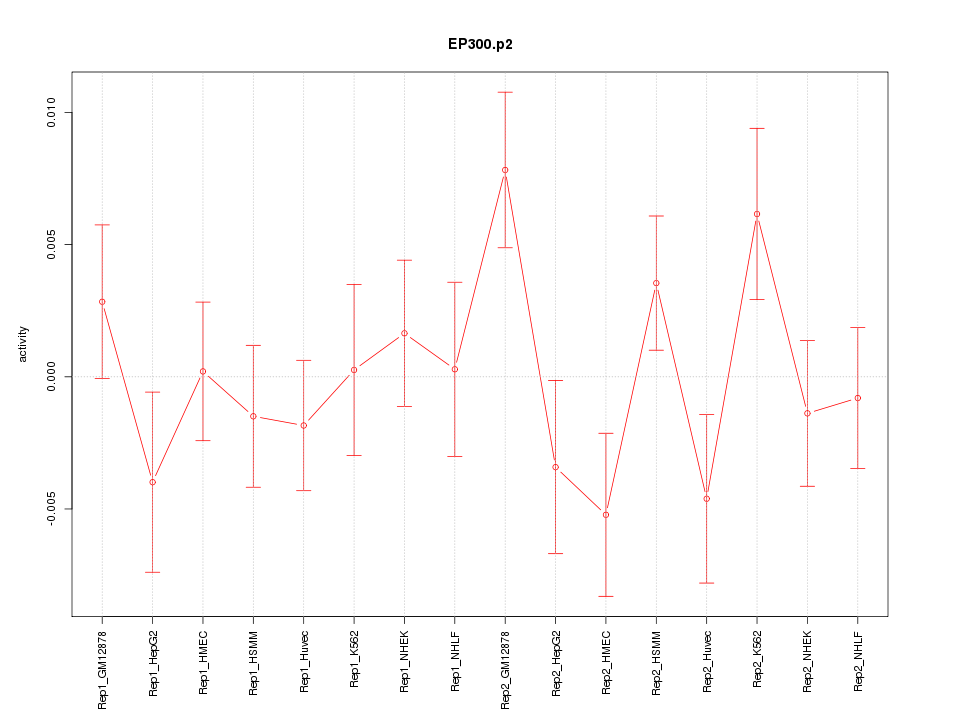
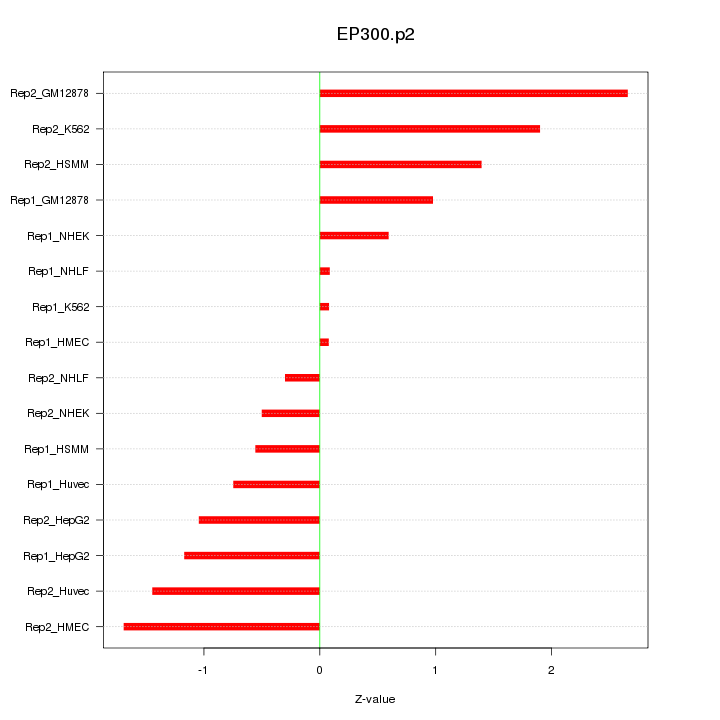
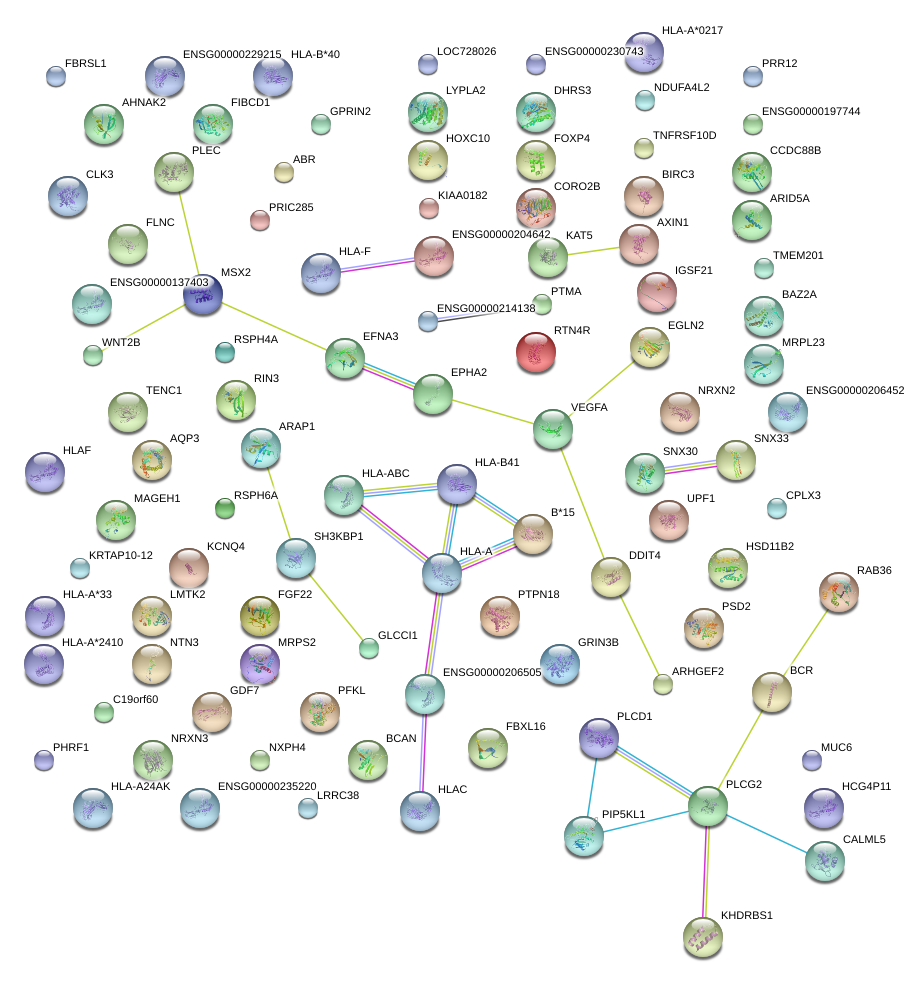
|
chr21_+_44941514
|
1.321
|
NM_198699
|
KRTAP10-12
|
keratin associated protein 10-12
|
|
chr9_-_132804057
|
1.292
|
NM_032843
|
FIBCD1
|
fibrinogen C domain containing 1
|
|
chr8_-_145085616
|
1.250
|
NM_201384
|
PLEC
|
plectin
|
|
chr9_-_132803809
|
1.188
|
|
FIBCD1
|
fibrinogen C domain containing 1
|
|
chr2_+_232281491
|
1.075
|
|
PTMA
|
prothymosin, alpha
|
|
chr9_+_114552871
|
1.046
|
NM_001012994
|
SNX30
|
sorting nexin family member 30
|
|
chr8_-_23077392
|
0.926
|
NM_003840
|
TNFRSF10D
|
tumor necrosis factor receptor superfamily, member 10d, decoy with truncated death domain
|
|
chr19_+_590878
|
0.907
|
NM_020637
|
FGF22
|
fibroblast growth factor 22
|
|
chr9_-_132804243
|
0.883
|
NM_001145106
|
FIBCD1
|
fibrinogen C domain containing 1
|
|
chr1_-_12599934
|
0.877
|
NM_004753
|
DHRS3
|
dehydrogenase/reductase (SDR family) member 3
|
|
chr16_+_84204424
|
0.851
|
NM_014615
|
KIAA0182
|
KIAA0182
|
|
chr9_-_129729518
|
0.845
|
NM_173492
|
PIP5KL1
|
phosphatidylinositol-4-phosphate 5-kinase-like 1
|
|
chr19_-_51010272
|
0.787
|
NM_030785
|
RSPH6A
|
radial spoke head 6 homolog A (Chlamydomonas)
|
|
chr6_+_43846735
|
0.779
|
|
VEGFA
|
vascular endothelial growth factor A
|
|
chr14_-_104515738
|
0.747
|
NM_138420
|
AHNAK2
|
AHNAK nucleoprotein 2
|
|
chr2_+_96566190
|
0.735
|
NM_212481
|
ARID5A
|
AT rich interactive domain 5A (MRF1-like)
|
|
chr7_+_7974941
|
0.727
|
NM_138426
|
GLCCI1
|
glucocorticoid induced transcript 1
|
|
chr7_+_128257698
|
0.726
|
NM_001127487
NM_001458
|
FLNC
|
filamin C, gamma
|
|
chr1_+_23990228
|
0.712
|
NM_007260
|
LYPLA2
|
lysophospholipase II
|
|
chr15_+_72695248
|
0.706
|
|
CLK3
|
CDC-like kinase 3
|
|
chr2_+_232281470
|
0.682
|
NM_001099285
NM_002823
|
PTMA
|
prothymosin, alpha
|
|
chr2_+_130846263
|
0.682
|
|
PTPN18
|
protein tyrosine phosphatase, non-receptor type 18 (brain-derived)
|
|
chr11_+_1925075
|
0.680
|
NM_021134
|
MRPL23
|
mitochondrial ribosomal protein L23
|
|
chr12_+_51727076
|
0.656
|
NM_198316
|
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2)
|
|
chrX_-_19598912
|
0.645
|
NM_001184960
|
SH3KBP1
|
SH3-domain kinase binding protein 1
|
|
chr10_-_5531401
|
0.643
|
NM_017422
|
CALML5
|
calmodulin-like 5
|
|
chr19_+_951295
|
0.632
|
NM_138690
|
GRIN3B
|
glutamate receptor, ionotropic, N-methyl-D-aspartate 3B
|
|
chr5_+_139155569
|
0.629
|
NM_032289
|
PSD2
|
pleckstrin and Sec7 domain containing 2
|
|
chr9_-_33437527
|
0.620
|
NM_004925
|
AQP3
|
aquaporin 3 (Gill blood group)
|
|
chr11_+_1925162
|
0.616
|
|
MRPL23
|
mitochondrial ribosomal protein L23
|
|
chr1_+_9571518
|
0.602
|
NM_001010866
NM_001130924
|
TMEM201
|
transmembrane protein 201
|
|
chr12_+_131577229
|
0.595
|
NM_001142641
|
FBRSL1
|
fibrosin-like 1
|
|
chr22_+_21852911
|
0.586
|
|
BCR
|
breakpoint cluster region
|
|
chr1_+_112852892
|
0.586
|
NM_024494
|
WNT2B
|
wingless-type MMTV integration site family, member 2B
|
|
chr16_-_342399
|
0.575
|
NM_003502
NM_181050
|
AXIN1
|
axin 1
|
|
chr12_+_52665196
|
0.572
|
NM_017409
|
HOXC10
|
homeobox C10
|
|
chr1_+_154878344
|
0.569
|
NM_021948
NM_198427
|
BCAN
|
brevican
|
|
chr20_-_61669550
|
0.567
|
NM_033405
|
PRIC285
|
peroxisomal proliferator-activated receptor A interacting complex 285
|
|
chr11_-_72111027
|
0.564
|
NM_001135190
NM_015242
|
ARAP1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1
|
|
chr15_+_72906003
|
0.560
|
NM_001030005
|
CPLX3
|
complexin 3
|
|
chr17_-_975809
|
0.559
|
|
ABR
|
active BCR-related gene
|
|
chr2_+_20729904
|
0.547
|
NM_182828
|
GDF7
|
growth differentiation factor 7
|
|
chr15_+_66658626
|
0.540
|
NM_006091
|
CORO2B
|
coronin, actin binding protein, 2B
|
|
chr22_+_21870293
|
0.531
|
|
|
|
|
chr22_+_21852699
|
0.528
|
|
BCR
|
breakpoint cluster region
|
|
chr3_-_38046034
|
0.524
|
NM_006225
|
PLCD1
|
phospholipase C, delta 1
|
|
chr6_+_30018287
|
0.524
|
NM_002116
|
HLA-A
|
major histocompatibility complex, class I, A
|
|
chr15_+_72687765
|
0.520
|
NM_003992
|
CLK3
|
CDC-like kinase 3
|
|
chr19_+_18803742
|
0.520
|
NM_002911
|
UPF1
|
UPF1 regulator of nonsense transcripts homolog (yeast)
|
|
chr10_+_73703682
|
0.513
|
NM_019058
|
DDIT4
|
DNA-damage-inducible transcript 4
|
|
chr22_+_21852743
|
0.510
|
|
BCR
|
breakpoint cluster region
|
|
chr14_+_92049396
|
0.498
|
NM_024832
|
RIN3
|
Ras and Rab interactor 3
|
|
chr11_+_65236261
|
0.496
|
|
KAT5
|
K(lysine) acetyltransferase 5
|
|
chr1_+_41022066
|
0.496
|
NM_004700
NM_172163
|
KCNQ4
|
potassium voltage-gated channel, KQT-like subfamily, member 4
|
|
chr10_+_46413551
|
0.493
|
NM_014696
|
GPRIN2
|
G protein regulated inducer of neurite outgrowth 2
|
|
chr22_-_18635614
|
0.488
|
NM_023004
|
RTN4R
|
reticulon 4 receptor
|
|
chr6_+_29799095
|
0.487
|
NM_001098478
NM_001098479
NM_018950
|
HLA-F
|
major histocompatibility complex, class I, F
|
|
chr6_+_41622267
|
0.485
|
|
FOXP4
|
forkhead box P4
|
|
chr19_+_18803786
|
0.484
|
|
UPF1
|
UPF1 regulator of nonsense transcripts homolog (yeast)
|
|
chr1_+_18306823
|
0.482
|
NM_032880
|
IGSF21
|
immunoglobin superfamily, member 21
|
|
chrX_+_55495262
|
0.477
|
NM_014061
|
MAGEH1
|
melanoma antigen family H, 1
|
|
chr21_+_44569537
|
0.476
|
|
PFKL
|
phosphofructokinase, liver
|
|
chr11_-_64247230
|
0.471
|
NM_015080
NM_138732
|
NRXN2
|
neurexin 2
|
|
chr22_+_21852551
|
0.467
|
NM_004327
NM_021574
|
BCR
|
breakpoint cluster region
|
|
chr11_+_566482
|
0.463
|
NM_020901
|
PHRF1
|
PHD and ring finger domains 1
|
|
chr5_+_174084137
|
0.463
|
NM_002449
|
MSX2
|
msh homeobox 2
|
|
chr11_+_101693403
|
0.462
|
NM_001165
NM_182962
|
BIRC3
|
baculoviral IAP repeat containing 3
|
|
chr19_+_54786723
|
0.462
|
NM_020719
|
PRR12
|
proline rich 12
|
|
chr12_-_55310261
|
0.461
|
|
BAZ2A
|
bromodomain adjacent to zinc finger domain, 2A
|
|
chr1_-_154205675
|
0.460
|
|
ARHGEF2
|
Rho/Rac guanine nucleotide exchange factor (GEF) 2
|
|
chr16_+_66022729
|
0.455
|
|
HSD11B2
|
hydroxysteroid (11-beta) dehydrogenase 2
|
|
chr16_-_695719
|
0.449
|
NM_153350
|
FBXL16
|
F-box and leucine-rich repeat protein 16
|
|
chr11_-_1007140
|
0.444
|
|
MUC6
|
mucin 6, oligomeric mucus/gel-forming
|
|
chr16_+_2461500
|
0.438
|
NM_006181
|
NTN3
|
netrin 3
|
|
chr12_+_55896844
|
0.435
|
NM_007224
|
NXPH4
|
neurexophilin 4
|
|
chr16_+_80370426
|
0.435
|
NM_002661
|
PLCG2
|
phospholipase C, gamma 2 (phosphatidylinositol-specific)
|
|
chr22_+_21817512
|
0.433
|
NM_004914
|
RAB36
|
RAB36, member RAS oncogene family
|
|
chr11_+_63864263
|
0.429
|
NM_032251
|
CCDC88B
|
coiled-coil domain containing 88B
|
|
chr1_-_13712744
|
0.427
|
NM_001010847
|
LRRC38
|
leucine rich repeat containing 38
|
|
chr19_+_18560494
|
0.423
|
NM_001100418
NM_001100419
|
C19orf60
|
chromosome 19 open reading frame 60
|
|
chr7_+_97574054
|
0.423
|
NM_014916
|
LMTK2
|
lemur tyrosine kinase 2
|
|
chr1_-_16355013
|
0.422
|
NM_004431
|
EPHA2
|
EPH receptor A2
|
|
chr12_-_55920741
|
0.417
|
NM_020142
|
NDUFA4L2
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4-like 2
|
|
chr1_+_153317887
|
0.409
|
NM_004952
|
EFNA3
|
ephrin-A3
|
|
chr1_+_32251988
|
0.403
|
NM_006559
|
KHDRBS1
|
KH domain containing, RNA binding, signal transduction associated 1
|
|
chr19_+_45998020
|
0.402
|
NM_053046
|
EGLN2
|
egl nine homolog 2 (C. elegans)
|
|
chr9_+_137532294
|
0.397
|
NM_016034
|
MRPS2
|
mitochondrial ribosomal protein S2
|
|
chr17_+_24095135
|
0.390
|
NM_004295
|
TRAF4
|
TNF receptor-associated factor 4
|
|
chr10_-_70846573
|
0.390
|
NM_001057
|
TACR2
|
tachykinin receptor 2
|
|
chr22_-_43040063
|
0.388
|
NM_001099294
|
KIAA1644
|
KIAA1644
|
|
chr3_-_51971915
|
0.387
|
NM_033010
|
PCBP4
|
poly(rC) binding protein 4
|
|
chr8_-_134156437
|
0.384
|
|
SLA
|
Src-like-adaptor
|
|
chr7_+_45163891
|
0.382
|
NM_005856
|
RAMP3
|
receptor (G protein-coupled) activity modifying protein 3
|
|
chrX_+_48529905
|
0.381
|
NM_002049
|
GATA1
|
GATA binding protein 1 (globin transcription factor 1)
|
|
chr6_-_33822659
|
0.380
|
NM_001142883
NM_054111
|
IP6K3
|
inositol hexakisphosphate kinase 3
|
|
chr7_-_142293382
|
0.380
|
NM_018646
|
TRPV6
|
transient receptor potential cation channel, subfamily V, member 6
|
|
chr1_+_24755152
|
0.380
|
NM_001010980
|
C1orf130
|
chromosome 1 open reading frame 130
|
|
chr16_+_2023279
|
0.377
|
|
SLC9A3R2
|
solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 2
|
|
chr10_-_80875097
|
0.376
|
NM_153367
|
ZCCHC24
|
zinc finger, CCHC domain containing 24
|
|
chr8_-_142307854
|
0.376
|
NM_001080431
|
SLC45A4
|
solute carrier family 45, member 4
|
|
chr10_-_49483002
|
0.373
|
NM_021226
|
ARHGAP22
|
Rho GTPase activating protein 22
|
|
chrX_-_131089552
|
0.372
|
NM_194277
|
FRMD7
|
FERM domain containing 7
|
|
chr19_+_52451642
|
0.371
|
|
CCDC9
|
coiled-coil domain containing 9
|
|
chr12_+_52806103
|
0.369
|
|
LOC400043
|
hypothetical LOC400043
|
|
chr20_+_34635362
|
0.369
|
NM_021809
|
TGIF2
|
TGFB-induced factor homeobox 2
|
|
chr17_-_24253821
|
0.365
|
NM_144683
|
DHRS13
|
dehydrogenase/reductase (SDR family) member 13
|
|
chr19_+_18391423
|
0.360
|
|
|
|
|
chr19_+_1358664
|
0.357
|
|
DAZAP1
|
DAZ associated protein 1
|
|
chr19_+_41039649
|
0.355
|
NM_032123
NM_199179
NM_199180
|
KIRREL2
|
kin of IRRE like 2 (Drosophila)
|
|
chr6_+_111409933
|
0.355
|
NM_032194
|
RPF2
|
ribosome production factor 2 homolog (S. cerevisiae)
|
|
chr11_+_47386621
|
0.355
|
NM_001128225
NM_152264
|
SLC39A13
|
solute carrier family 39 (zinc transporter), member 13
|
|
chr5_+_133478252
|
0.354
|
NM_003202
|
TCF7
|
transcription factor 7 (T-cell specific, HMG-box)
|
|
chr1_+_32346206
|
0.354
|
NM_012316
|
KPNA6
|
karyopherin alpha 6 (importin alpha 7)
|
|
chr9_+_126671321
|
0.353
|
|
ARPC5L
|
actin related protein 2/3 complex, subunit 5-like
|
|
chr1_+_36327039
|
0.351
|
NM_017825
|
ADPRHL2
|
ADP-ribosylhydrolase like 2
|
|
chr14_+_105028614
|
0.350
|
NM_001134875
NM_001134876
|
C14orf80
|
chromosome 14 open reading frame 80
|
|
chr19_+_1358531
|
0.349
|
NM_018959
NM_170711
|
DAZAP1
|
DAZ associated protein 1
|
|
chr1_+_179148935
|
0.347
|
NM_020950
|
KIAA1614
|
KIAA1614
|
|
chr20_+_43468046
|
0.345
|
NM_001197139
NM_001048221
NM_001048222
NM_001197140
NM_018478
|
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2
|
|
chr1_-_23567084
|
0.342
|
NM_030634
|
ZNF436
|
zinc finger protein 436
|
|
chr3_+_48482625
|
0.341
|
|
TREX1
|
three prime repair exonuclease 1
|
|
chrX_+_67965517
|
0.341
|
NM_004429
|
EFNB1
|
ephrin-B1
|
|
chr1_+_151917674
|
0.340
|
NM_000906
|
NPR1
|
natriuretic peptide receptor A/guanylate cyclase A (atrionatriuretic peptide receptor A)
|
|
chr7_+_74877568
|
0.339
|
|
NSUN5P1
|
NOP2/Sun domain family, member 5 pseudogene 1
|
|
chr1_-_2451543
|
0.339
|
NM_001010926
|
HES5
|
hairy and enhancer of split 5 (Drosophila)
|
|
chr8_-_134156353
|
0.338
|
|
SLA
|
Src-like-adaptor
|
|
chr15_+_62921134
|
0.337
|
NM_001195059
NM_025201
|
PLEKHO2
|
pleckstrin homology domain containing, family O member 2
|
|
chr11_+_46679140
|
0.337
|
NM_001184751
|
ZNF408
|
zinc finger protein 408
|
|
chr3_+_46898673
|
0.335
|
NM_001184744
|
PTH1R
|
parathyroid hormone 1 receptor
|
|
chr9_+_139152595
|
0.334
|
|
GRIN1
|
glutamate receptor, ionotropic, N-methyl D-aspartate 1
|
|
chr19_+_1358604
|
0.333
|
|
DAZAP1
|
DAZ associated protein 1
|
|
chr5_+_176786279
|
0.333
|
NM_001004105
NM_001004106
NM_002082
|
GRK6
|
G protein-coupled receptor kinase 6
|
|
chr22_-_30356783
|
0.332
|
NM_014338
|
PISD
|
phosphatidylserine decarboxylase
|
|
chr20_-_29775354
|
0.332
|
|
BCL2L1
|
BCL2-like 1
|
|
chr22_+_18081968
|
0.331
|
NM_002688
|
SEPT5
|
septin 5
|
|
chr22_+_39404940
|
0.329
|
NM_005297
|
MCHR1
|
melanin-concentrating hormone receptor 1
|
|
chr11_-_6451561
|
0.328
|
NM_006458
NM_033278
|
TRIM3
|
tripartite motif containing 3
|
|
chr6_+_30565161
|
0.328
|
NM_005516
|
HLA-E
|
major histocompatibility complex, class I, E
|
|
chr19_+_12966970
|
0.326
|
NM_002501
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor)
|
|
chr11_+_66547391
|
0.325
|
NM_177963
|
SYT12
|
synaptotagmin XII
|
|
chr6_+_30018265
|
0.325
|
|
HLA-A
|
major histocompatibility complex, class I, A
|
|
chrX_+_69589081
|
0.322
|
|
DLG3
|
discs, large homolog 3 (Drosophila)
|
|
chr10_+_134060681
|
0.320
|
NM_001098637
NM_138499
|
PWWP2B
|
PWWP domain containing 2B
|
|
chr16_-_85099941
|
0.319
|
|
LOC400550
|
hypothetical LOC400550
|
|
chr19_+_10900502
|
0.317
|
|
C19orf52
|
chromosome 19 open reading frame 52
|
|
chr16_-_30274182
|
0.316
|
NM_006110
|
CD2BP2
|
CD2 (cytoplasmic tail) binding protein 2
|
|
chr6_-_31890765
|
0.316
|
NM_005527
|
HSPA1L
|
heat shock 70kDa protein 1-like
|
|
chrX_-_17789276
|
0.316
|
NM_001172732
NM_001172739
NM_001172743
NM_021785
|
RAI2
|
retinoic acid induced 2
|
|
chr21_+_41655786
|
0.315
|
NM_002463
|
LOC100131190
MX2
|
hypothetical protein LOC100131190
myxovirus (influenza virus) resistance 2 (mouse)
|
|
chr15_+_62539831
|
0.315
|
|
ZNF609
|
zinc finger protein 609
|
|
chr12_+_52653176
|
0.314
|
NM_014212
|
HOXC11
|
homeobox C11
|
|
chr7_-_155294518
|
0.313
|
|
SHH
|
sonic hedgehog
|
|
chr1_+_22176004
|
0.313
|
NM_007352
|
CELA3A
CELA3B
|
chymotrypsin-like elastase family, member 3A
chymotrypsin-like elastase family, member 3B
|
|
chr17_+_35075124
|
0.312
|
NM_003673
|
TCAP
|
titin-cap (telethonin)
|
|
chr17_+_35751796
|
0.311
|
NM_001024809
|
RARA
|
retinoic acid receptor, alpha
|
|
chrX_+_46977306
|
0.310
|
|
USP11
|
ubiquitin specific peptidase 11
|
|
chr12_+_48652886
|
0.310
|
NM_001652
|
AQP6
|
aquaporin 6, kidney specific
|
|
chrX_+_46977208
|
0.306
|
NM_004651
|
USP11
|
ubiquitin specific peptidase 11
|
|
chr22_-_49025499
|
0.306
|
NM_020461
|
TUBGCP6
|
tubulin, gamma complex associated protein 6
|
|
chr11_+_56984702
|
0.305
|
NM_178570
|
RTN4RL2
|
reticulon 4 receptor-like 2
|
|
chr12_-_122684150
|
0.305
|
NM_001414
|
EIF2B1
|
eukaryotic translation initiation factor 2B, subunit 1 alpha, 26kDa
|
|
chrX_-_151670754
|
0.305
|
NM_153488
|
MAGEA2B
MAGEA2
|
melanoma antigen family A, 2B
melanoma antigen family A, 2
|
|
chr6_-_31756119
|
0.305
|
NM_025262
|
LY6G5C
|
lymphocyte antigen 6 complex, locus G5C
|
|
chr12_-_113606322
|
0.304
|
NM_005996
NM_016569
|
TBX3
|
T-box 3
|
|
chr22_+_22907417
|
0.304
|
|
SUSD2
|
sushi domain containing 2
|
|
chr7_-_100331417
|
0.303
|
NM_000665
NM_015831
|
ACHE
|
acetylcholinesterase
|
|
chr9_-_116920232
|
0.303
|
NM_002160
|
TNC
|
tenascin C
|
|
chr22_-_20637188
|
0.302
|
|
PPM1F
|
protein phosphatase, Mg2+/Mn2+ dependent, 1F
|
|
chr1_-_159275357
|
0.302
|
NM_001113205
NM_001113206
NM_001113207
|
TSTD1
F11R
|
thiosulfate sulfurtransferase (rhodanese)-like domain containing 1
F11 receptor
|
|
chr1_+_199519202
|
0.300
|
NM_000299
NM_001005337
|
PKP1
|
plakophilin 1 (ectodermal dysplasia/skin fragility syndrome)
|
|
chr15_+_72892205
|
0.300
|
NM_021819
|
LMAN1L
|
lectin, mannose-binding, 1 like
|
|
chr22_-_20637212
|
0.300
|
NM_014634
|
PPM1F
|
protein phosphatase, Mg2+/Mn2+ dependent, 1F
|
|
chr11_+_67563093
|
0.299
|
|
TCIRG1
|
T-cell, immune regulator 1, ATPase, H+ transporting, lysosomal V0 subunit A3
|
|
chr22_+_22907443
|
0.298
|
NM_019601
|
SUSD2
|
sushi domain containing 2
|
|
chrX_+_140819307
|
0.298
|
NM_005462
|
MAGEC1
|
melanoma antigen family C, 1
|
|
chr12_+_115833093
|
0.298
|
NM_012174
NM_153348
|
FBXW8
|
F-box and WD repeat domain containing 8
|
|
chr17_+_2646481
|
0.297
|
NM_001100398
NM_015085
|
RAP1GAP2
|
RAP1 GTPase activating protein 2
|
|
chr11_+_133599771
|
0.297
|
|
VPS26B
|
vacuolar protein sorting 26 homolog B (S. pombe)
|
|
chr18_+_30810889
|
0.296
|
NM_001143826
|
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2
|
|
chr16_+_29892714
|
0.295
|
NM_004783
NM_016151
|
TAOK2
|
TAO kinase 2
|
|
chr22_-_27405708
|
0.295
|
NM_001145418
|
TTC28
|
tetratricopeptide repeat domain 28
|
|
chr1_-_159282312
|
0.294
|
|
USF1
|
upstream transcription factor 1
|
|
chr22_+_20251956
|
0.294
|
NM_003347
|
UBE2L3
|
ubiquitin-conjugating enzyme E2L 3
|
|
chr14_-_87859284
|
0.293
|
NM_138317
|
KCNK10
|
potassium channel, subfamily K, member 10
|
|
chr16_+_2145765
|
0.292
|
NM_032271
|
TRAF7
|
TNF receptor-associated factor 7
|
|
chr11_+_133599755
|
0.291
|
NM_052875
|
VPS26B
|
vacuolar protein sorting 26 homolog B (S. pombe)
|
|
chr22_-_20314274
|
0.291
|
NM_001017964
|
YDJC
|
YdjC homolog (bacterial)
|
|
chr11_+_133599689
|
0.289
|
|
VPS26B
|
vacuolar protein sorting 26 homolog B (S. pombe)
|
|
chr12_+_122684265
|
0.289
|
NM_001516
|
GTF2H3
|
general transcription factor IIH, polypeptide 3, 34kDa
|
|
chr15_+_88345612
|
0.289
|
NM_198526
|
ZNF710
|
zinc finger protein 710
|
|
chr1_+_203064397
|
0.288
|
NM_001005388
NM_001005389
NM_001160332
NM_001160333
NM_015090
|
NFASC
|
neurofascin
|
|
chr11_-_133599628
|
0.287
|
NM_015261
|
NCAPD3
|
non-SMC condensin II complex, subunit D3
|
|
chr3_+_128874458
|
0.286
|
NM_172027
|
ABTB1
|
ankyrin repeat and BTB (POZ) domain containing 1
|
|
chr22_+_18330049
|
0.286
|
NM_007310
|
COMT
|
catechol-O-methyltransferase
|
|
chr11_+_47386707
|
0.286
|
|
SLC39A13
|
solute carrier family 39 (zinc transporter), member 13
|
|
chr3_+_157874898
|
0.286
|
NM_015508
|
TIPARP
|
TCDD-inducible poly(ADP-ribose) polymerase
|
|
chr19_-_18749728
|
0.286
|
|
|
|
|
chr16_-_960796
|
0.283
|
|
LMF1
|
lipase maturation factor 1
|
|
chr22_+_20252004
|
0.281
|
|
UBE2L3
|
ubiquitin-conjugating enzyme E2L 3
|
|
chr9_-_131555111
|
0.280
|
NM_004878
|
PTGES
|
prostaglandin E synthase
|