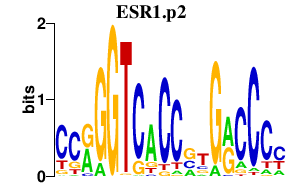
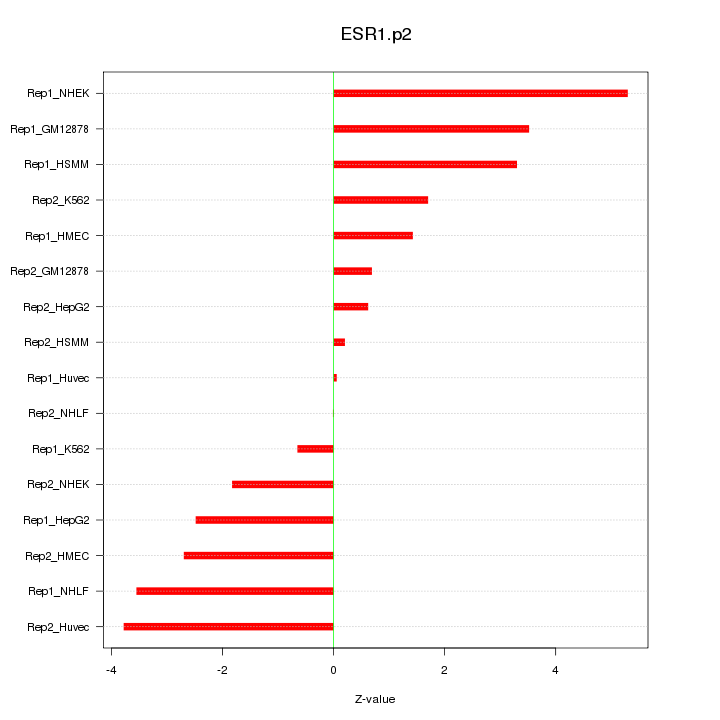
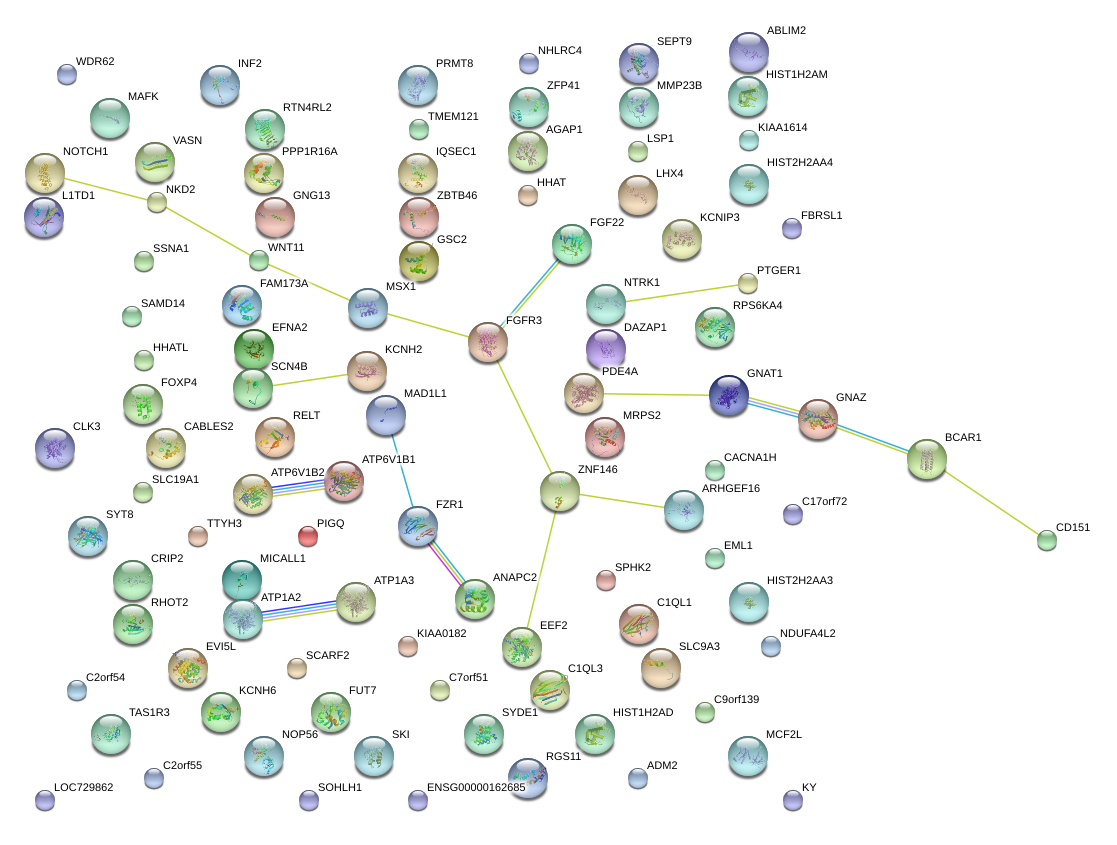
|
chr19_+_1236766
|
4.270
|
NM_001405
|
EFNA2
|
ephrin-A2
|
|
chr21_-_45786709
|
3.351
|
NM_194255
|
SLC19A1
|
solute carrier family 19 (folate transporter), member 1
|
|
chr16_+_550394
|
2.604
|
NM_145270
|
C16orf11
|
chromosome 16 open reading frame 11
|
|
chr16_+_559970
|
2.524
|
NM_004204
NM_148920
|
PIGQ
|
phosphatidylinositol glycan anchor biosynthesis, class Q
|
|
chr19_-_3927523
|
2.482
|
|
EEF2
|
eukaryotic translation elongation factor 2
|
|
chr3_-_135852514
|
2.432
|
NM_178554
|
KY
|
kyphoscoliosis peptidase
|
|
chr16_+_84204424
|
2.288
|
NM_014615
|
KIAA0182
|
KIAA0182
|
|
chr1_+_178466064
|
2.242
|
NM_033343
|
LHX4
|
LIM homeobox 4
|
|
chr19_+_3457264
|
2.227
|
NM_016263
|
FZR1
|
fizzy/cell division cycle 20 related 1 (Drosophila)
|
|
chr9_+_139202874
|
2.212
|
NM_003731
|
SSNA1
|
Sjogren syndrome nuclear autoantigen 1
|
|
chr14_+_105063975
|
2.131
|
NM_025268
|
TMEM121
|
transmembrane protein 121
|
|
chr1_+_1256585
|
2.099
|
NM_152228
|
TAS1R3
|
taste receptor, type 1, member 3
|
|
chr9_+_137532294
|
2.046
|
NM_016034
|
MRPS2
|
mitochondrial ribosomal protein S2
|
|
chr8_+_144428083
|
2.036
|
|
GLI4
|
GLI family zinc finger 4
|
|
chr19_+_590878
|
2.027
|
NM_020637
|
FGF22
|
fibroblast growth factor 22
|
|
chr7_+_1536847
|
1.985
|
NM_002360
|
MAFK
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog K (avian)
|
|
chr20_-_60415729
|
1.977
|
NM_031215
|
CABLES2
|
Cdk5 and Abl enzyme substrate 2
|
|
chr19_+_10392128
|
1.920
|
NM_001111307
|
PDE4A
|
phosphodiesterase 4A, cAMP-specific
|
|
chr1_+_62433061
|
1.841
|
NM_001164835
NM_019079
|
L1TD1
|
LINE-1 type transposase domain containing 1
|
|
chr9_-_139202855
|
1.822
|
NM_013366
|
ANAPC2
|
anaphase promoting complex subunit 2
|
|
chr7_+_1543644
|
1.808
|
|
MAFK
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog K (avian)
|
|
chr1_+_1062137
|
1.808
|
|
LOC254099
|
hypothetical protein LOC254099
|
|
chr16_+_4361775
|
1.792
|
NM_138440
|
VASN
|
vasorin
|
|
chr19_-_14447163
|
1.785
|
NM_000955
|
PTGER1
|
prostaglandin E receptor 1 (subtype EP1), 42kDa
|
|
chr4_+_1764802
|
1.782
|
NM_000142
NM_001163213
NM_022965
|
FGFR3
|
fibroblast growth factor receptor 3
|
|
chr22_+_36632242
|
1.764
|
NM_033386
|
MICALL1
|
MICAL-like 1
|
|
chr19_+_3473953
|
1.754
|
NM_001136197
NM_001136198
|
FZR1
|
fizzy/cell division cycle 20 related 1 (Drosophila)
|
|
chr14_+_99273751
|
1.696
|
|
EML1
|
echinoderm microtubule associated protein like 1
|
|
chr17_-_45562112
|
1.686
|
NM_174920
|
SAMD14
|
sterile alpha motif domain containing 14
|
|
chr9_+_139047441
|
1.673
|
|
C9orf139
|
chromosome 9 open reading frame 139
|
|
chr8_+_145692916
|
1.672
|
NM_032902
|
PPP1R16A
|
protein phosphatase 1, regulatory (inhibitor) subunit 16A
|
|
chr9_-_139202831
|
1.667
|
|
ANAPC2
|
anaphase promoting complex subunit 2
|
|
chr1_+_3360880
|
1.665
|
NM_014448
|
ARHGEF16
|
Rho guanine nucleotide exchange factor (GEF) 16
|
|
chr19_-_14447122
|
1.652
|
|
PTGER1
|
prostaglandin E receptor 1 (subtype EP1), 42kDa
|
|
chr1_+_2149993
|
1.641
|
NM_003036
|
SKI
|
v-ski sarcoma viral oncogene homolog (avian)
|
|
chr9_-_139202804
|
1.631
|
|
ANAPC2
|
anaphase promoting complex subunit 2
|
|
chr19_+_15079141
|
1.566
|
NM_033025
|
SYDE1
|
synapse defective 1, Rho GTPase, homolog 1 (C. elegans)
|
|
chr11_+_63883192
|
1.546
|
NM_001006944
NM_003942
|
RPS6KA4
|
ribosomal protein S6 kinase, 90kDa, polypeptide 4
|
|
chr13_+_112789669
|
1.543
|
|
MCF2L
|
MCF.2 cell line derived transforming sequence-like
|
|
chr16_+_711139
|
1.541
|
NM_023933
|
FAM173A
|
family with sequence similarity 173, member A
|
|
chr17_-_40401138
|
1.528
|
NM_006688
|
C1QL1
|
complement component 1, q subcomponent-like 1
|
|
chr1_+_148089251
|
1.526
|
NM_001040874
NM_003516
|
HIST2H2AA4
HIST2H2AA3
|
histone cluster 2, H2aa4
histone cluster 2, H2aa3
|
|
chr14_+_104244998
|
1.500
|
|
INF2
|
inverted formin, FH2 and WH2 domain containing
|
|
chr17_+_59429442
|
1.495
|
NM_001164257
NM_001191029
NM_001191030
NM_001191031
|
C17orf72
|
chromosome 17 open reading frame 72
|
|
chr22_+_21742727
|
1.446
|
|
GNAZ
|
guanine nucleotide binding protein (G protein), alpha z polypeptide
|
|
chr1_+_1557419
|
1.442
|
NM_006983
|
MMP23B
|
matrix metallopeptidase 23B
|
|
chr22_+_49266836
|
1.438
|
NM_024866
|
ADM2
|
adrenomedullin 2
|
|
chr22_-_17517795
|
1.434
|
NM_005315
|
GSC2
|
goosecoid homeobox 2
|
|
chr5_-_577444
|
1.433
|
NM_004174
|
SLC9A3
|
solute carrier family 9 (sodium/hydrogen exchanger), member 3
|
|
chr11_+_1846478
|
1.425
|
NM_001013253
|
LSP1
|
lymphocyte-specific protein 1
|
|
chr19_+_41397343
|
1.415
|
NM_001099638
NM_001099639
NM_007145
|
ZNF146
|
zinc finger protein 146
|
|
chr6_+_41622111
|
1.400
|
NM_001012426
NM_001012427
NM_138457
|
FOXP4
|
forkhead box P4
|
|
chr9_-_139047112
|
1.398
|
NM_004479
|
FUT7
|
fucosyltransferase 7 (alpha (1,3) fucosyltransferase)
|
|
chr12_-_123568781
|
1.390
|
|
|
|
|
chr16_-_790699
|
1.385
|
NM_016541
|
GNG13
|
guanine nucleotide binding protein (G protein), gamma 13
|
|
chr3_-_13089547
|
1.380
|
NM_001134382
|
IQSEC1
|
IQ motif and Sec7 domain 1
|
|
chr22_+_21742364
|
1.369
|
NM_002073
|
GNAZ
|
guanine nucleotide binding protein (G protein), alpha z polypeptide
|
|
chr16_+_1143241
|
1.362
|
NM_001005407
NM_021098
|
CACNA1H
|
calcium channel, voltage-dependent, T type, alpha 1H subunit
|
|
chr19_+_53819460
|
1.360
|
|
SPHK2
|
sphingosine kinase 2
|
|
chr9_-_138560058
|
1.354
|
NM_017617
|
NOTCH1
|
notch 1
|
|
chr12_+_131577229
|
1.353
|
NM_001142641
|
FBRSL1
|
fibrosin-like 1
|
|
chr2_+_95326753
|
1.339
|
NM_013434
|
KCNIP3
|
Kv channel interacting protein 3, calsenilin
|
|
chr15_+_72695248
|
1.338
|
|
CLK3
|
CDC-like kinase 3
|
|
chr20_-_61933012
|
1.336
|
|
ZBTB46
|
zinc finger and BTB domain containing 46
|
|
chr11_+_822965
|
1.298
|
|
CD151
|
CD151 molecule (Raph blood group)
|
|
chr19_-_50963942
|
1.294
|
|
|
|
|
chr11_+_1812115
|
1.291
|
NM_138567
|
SYT8
|
synaptotagmin VIII
|
|
chr11_+_1830763
|
1.279
|
NM_002339
|
LSP1
|
lymphocyte-specific protein 1
|
|
chr16_-_265868
|
1.278
|
NM_003834
NM_183337
|
RGS11
|
regulator of G-protein signaling 11
|
|
chr19_-_47190160
|
1.260
|
NM_152296
|
ATP1A3
|
ATPase, Na+/K+ transporting, alpha 3 polypeptide
|
|
chr3_-_42719322
|
1.245
|
NM_020707
|
HHATL
|
hedgehog acyltransferase-like
|
|
chr19_+_7834124
|
1.227
|
|
EVI5L
|
ecotropic viral integration site 5-like
|
|
chr3_+_50204026
|
1.218
|
NM_000172
NM_144499
|
GNAT1
|
guanine nucleotide binding protein (G protein), alpha transducing activity polypeptide 1
|
|
chr9_-_137731139
|
1.215
|
NM_001012415
NM_001101677
|
SOHLH1
|
spermatogenesis and oogenesis specific basic helix-loop-helix 1
|
|
chr19_-_47190061
|
1.210
|
|
ATP1A3
|
ATPase, Na+/K+ transporting, alpha 3 polypeptide
|
|
chr1_+_225982862
|
1.192
|
|
LOC100130093
|
hypothetical LOC100130093
|
|
chr16_+_658081
|
1.192
|
NM_138769
|
RHOT2
|
ras homolog gene family, member T2
|
|
chr1_+_179148935
|
1.191
|
NM_020950
|
KIAA1614
|
KIAA1614
|
|
chr11_+_56984702
|
1.190
|
NM_178570
|
RTN4RL2
|
reticulon 4 receptor-like 2
|
|
chr5_+_1061900
|
1.187
|
NM_033120
|
NKD2
|
naked cuticle homolog 2 (Drosophila)
|
|
chr13_+_112681626
|
1.187
|
NM_024979
|
MCF2L
|
MCF.2 cell line derived transforming sequence-like
|
|
chr1_+_155097230
|
1.185
|
NM_001012331
NM_002529
|
NTRK1
|
neurotrophic tyrosine kinase, receptor, type 1
|
|
chr19_+_1358750
|
1.184
|
|
DAZAP1
|
DAZ associated protein 1
|
|
chr7_-_150283794
|
1.178
|
NM_172057
|
KCNH2
|
potassium voltage-gated channel, subfamily H (eag-related), member 2
|
|
chr9_-_101621964
|
1.178
|
|
|
|
|
chr7_+_2638156
|
1.176
|
|
TTYH3
|
tweety homolog 3 (Drosophila)
|
|
chr2_-_241480058
|
1.173
|
NM_024861
|
C2orf54
|
chromosome 2 open reading frame 54
|
|
chr12_+_3470685
|
1.171
|
NM_019854
|
PRMT8
|
protein arginine methyltransferase 8
|
|
chr2_+_236067424
|
1.165
|
NM_001037131
NM_014914
|
AGAP1
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 1
|
|
chr12_-_55920741
|
1.155
|
NM_020142
|
NDUFA4L2
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4-like 2
|
|
chr19_+_1358531
|
1.147
|
NM_018959
NM_170711
|
DAZAP1
|
DAZ associated protein 1
|
|
chr4_-_8124611
|
1.142
|
|
ABLIM2
|
actin binding LIM protein family, member 2
|
|
chr1_+_155157047
|
1.141
|
NM_144702
|
LRRC71
|
leucine rich repeat containing 71
|
|
chr11_-_117528744
|
1.139
|
NM_001142348
NM_174934
|
SCN4B
|
sodium channel, voltage-gated, type IV, beta
|
|
chr11_+_72765318
|
1.137
|
NM_032871
|
RELT
|
RELT tumor necrosis factor receptor
|
|
chr2_-_98919109
|
1.134
|
NM_207362
|
C2orf55
|
chromosome 2 open reading frame 55
|
|
chr7_-_1946667
|
1.133
|
|
MAD1L1
|
MAD1 mitotic arrest deficient-like 1 (yeast)
|
|
chr22_-_19122047
|
1.129
|
|
SCARF2
|
scavenger receptor class F, member 2
|
|
chr2_+_71016503
|
1.114
|
NM_001692
|
ATP6V1B1
|
ATPase, H+ transporting, lysosomal 56/58kDa, V1 subunit B1
|
|
chr11_-_75599433
|
1.108
|
|
WNT11
|
wingless-type MMTV integration site family, member 11
|
|
chr22_+_17539198
|
1.107
|
|
|
|
|
chr4_+_4912272
|
1.105
|
NM_002448
|
MSX1
|
msh homeobox 1
|
|
chr16_-_73842884
|
1.099
|
NM_001170717
NM_014567
|
BCAR1
|
breast cancer anti-estrogen resistance 1
|
|
chr16_+_658158
|
1.094
|
|
RHOT2
|
ras homolog gene family, member T2
|
|
chr7_+_99919485
|
1.088
|
NM_173564
|
C7orf51
|
chromosome 7 open reading frame 51
|
|
chr14_+_105012106
|
1.084
|
NM_001312
|
CRIP2
|
cysteine-rich protein 2
|
|
chr20_+_2581177
|
1.083
|
NM_006392
|
NOP56
|
NOP56 ribonucleoprotein homolog (yeast)
|
|
chr1_+_19864366
|
1.078
|
NM_000871
|
HTR6
|
5-hydroxytryptamine (serotonin) receptor 6
|
|
chr8_-_145090892
|
1.073
|
NM_201381
|
PLEC
|
plectin
|
|
chr11_-_69228286
|
1.067
|
NM_005117
|
FGF19
|
fibroblast growth factor 19
|
|
chr10_+_102881050
|
1.066
|
NM_001195517
NM_005521
|
TLX1
|
T-cell leukemia homeobox 1
|
|
chr17_+_77261457
|
1.065
|
|
|
|
|
chr10_-_133971136
|
1.064
|
NM_173575
|
STK32C
|
serine/threonine kinase 32C
|
|
chr3_-_197201497
|
1.063
|
|
SDHAP1
SDHAP2
|
succinate dehydrogenase complex, subunit A, flavoprotein pseudogene 1
succinate dehydrogenase complex, subunit A, flavoprotein pseudogene 2
|
|
chr6_+_36346214
|
1.053
|
NM_001145717
|
PNPLA1
|
patatin-like phospholipase domain containing 1
|
|
chr3_-_129690056
|
1.049
|
NM_001145661
|
GATA2
|
GATA binding protein 2
|
|
chr2_+_242398832
|
1.036
|
NM_001167600
NM_001167601
|
NEU4
|
sialidase 4
|
|
chr1_-_229070796
|
1.035
|
NM_032800
|
C1orf198
|
chromosome 1 open reading frame 198
|
|
chr19_+_1389336
|
1.033
|
NM_001018
|
RPS15
|
ribosomal protein S15
|
|
chr10_+_94811010
|
1.030
|
NM_183374
|
CYP26C1
|
cytochrome P450, family 26, subfamily C, polypeptide 1
|
|
chr17_+_4972410
|
1.027
|
NM_004505
|
USP6
|
ubiquitin specific peptidase 6 (Tre-2 oncogene)
|
|
chr11_-_72031083
|
1.026
|
NM_001143839
|
PDE2A
|
phosphodiesterase 2A, cGMP-stimulated
|
|
chr17_-_6400470
|
1.017
|
NM_001165966
NM_031220
|
PITPNM3
|
PITPNM family member 3
|
|
chr16_+_751050
|
1.006
|
NM_013404
|
MSLN
|
mesothelin
|
|
chr16_+_666075
|
1.002
|
NM_003961
|
RHBDL1
|
rhomboid, veinlet-like 1 (Drosophila)
|
|
chr19_-_2187246
|
1.002
|
NM_018049
|
PLEKHJ1
|
pleckstrin homology domain containing, family J member 1
|
|
chr19_-_41397822
|
1.001
|
NM_001042474
|
ZNF565
|
zinc finger protein 565
|
|
chr17_-_77462325
|
0.999
|
|
PCYT2
|
phosphate cytidylyltransferase 2, ethanolamine
|
|
chr19_+_3884100
|
0.999
|
NM_170678
|
ITGB1BP3
|
integrin beta 1 binding protein 3
|
|
chr2_+_242398954
|
0.995
|
|
NEU4
|
sialidase 4
|
|
chr11_-_1549719
|
0.993
|
NM_004420
|
DUSP8
|
dual specificity phosphatase 8
|
|
chr22_-_19122107
|
0.989
|
NM_153334
NM_182895
|
SCARF2
|
scavenger receptor class F, member 2
|
|
chr22_+_41836697
|
0.987
|
NM_001197
|
BIK
|
BCL2-interacting killer (apoptosis-inducing)
|
|
chr19_+_1389397
|
0.983
|
|
RPS15
|
ribosomal protein S15
|
|
chr5_-_172594691
|
0.982
|
NM_001166175
NM_001166176
NM_004387
|
NKX2-5
|
NK2 transcription factor related, locus 5 (Drosophila)
|
|
chr15_+_66658626
|
0.981
|
NM_006091
|
CORO2B
|
coronin, actin binding protein, 2B
|
|
chr17_+_77261440
|
0.972
|
|
HGS
|
hepatocyte growth factor-regulated tyrosine kinase substrate
|
|
chr16_-_87535106
|
0.970
|
NM_175931
|
CBFA2T3
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 3
|
|
chr1_-_13712744
|
0.967
|
NM_001010847
|
LRRC38
|
leucine rich repeat containing 38
|
|
chr16_+_87765661
|
0.964
|
NM_004933
|
CDH15
|
cadherin 15, type 1, M-cadherin (myotubule)
|
|
chr11_+_71613472
|
0.964
|
NM_001567
|
INPPL1
|
inositol polyphosphate phosphatase-like 1
|
|
chr19_+_10388448
|
0.957
|
|
PDE4A
|
phosphodiesterase 4A, cAMP-specific
|
|
chr17_+_77261477
|
0.954
|
|
HGS
|
hepatocyte growth factor-regulated tyrosine kinase substrate
|
|
chr11_+_61032847
|
0.950
|
NM_001145077
|
LRRC10B
|
leucine rich repeat containing 10B
|
|
chr20_+_773283
|
0.941
|
NM_031424
|
FAM110A
|
family with sequence similarity 110, member A
|
|
chr20_-_29896981
|
0.932
|
NM_004118
|
FOXS1
|
forkhead box S1
|
|
chr20_+_54637714
|
0.927
|
NM_003222
|
TFAP2C
|
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma)
|
|
chr10_+_112247614
|
0.926
|
NM_004419
|
DUSP5
|
dual specificity phosphatase 5
|
|
chr17_+_77261401
|
0.925
|
NM_004712
|
HGS
|
hepatocyte growth factor-regulated tyrosine kinase substrate
|
|
chr4_+_2013517
|
0.922
|
NM_001168243
NM_001141936
|
C4orf48
|
chromosome 4 open reading frame 48
|
|
chr17_+_77453363
|
0.921
|
NM_148896
|
NPB
|
neuropeptide B
|
|
chr16_-_3567354
|
0.919
|
NM_178844
|
NLRC3
|
NLR family, CARD domain containing 3
|
|
chr9_-_85342868
|
0.914
|
NM_174938
|
FRMD3
|
FERM domain containing 3
|
|
chr19_+_7874701
|
0.912
|
|
MAP2K7
|
mitogen-activated protein kinase kinase 7
|
|
chr19_+_16544360
|
0.912
|
|
|
|
|
chr21_-_44485269
|
0.908
|
|
ICOSLG
|
inducible T-cell co-stimulator ligand
|
|
chr8_-_142586487
|
0.907
|
NM_207414
|
FLJ43860
|
FLJ43860 protein
|
|
chr9_-_139060416
|
0.894
|
NM_015392
|
NPDC1
|
neural proliferation, differentiation and control, 1
|
|
chr11_+_65100074
|
0.894
|
NM_001099409
|
EHBP1L1
|
EH domain binding protein 1-like 1
|
|
chr19_+_4423149
|
0.893
|
NM_001001520
NM_032631
|
HDGFRP2
|
hepatoma-derived growth factor-related protein 2
|
|
chr17_+_77261447
|
0.893
|
|
HGS
|
hepatocyte growth factor-regulated tyrosine kinase substrate
|
|
chr1_+_151655623
|
0.883
|
NM_176823
|
S100A7A
|
S100 calcium binding protein A7A
|
|
chr6_+_30002084
|
0.879
|
|
|
|
|
chr19_+_7874764
|
0.877
|
NM_145185
|
MAP2K7
|
mitogen-activated protein kinase kinase 7
|
|
chr7_-_4965270
|
0.872
|
NM_001100600
NM_198403
|
MMD2
|
monocyte to macrophage differentiation-associated 2
|
|
chr5_-_1165106
|
0.871
|
NM_006598
|
SLC12A7
|
solute carrier family 12 (potassium/chloride transporters), member 7
|
|
chr17_-_77462506
|
0.869
|
NM_001184917
NM_002861
|
PCYT2
|
phosphate cytidylyltransferase 2, ethanolamine
|
|
chr10_-_103525646
|
0.867
|
NM_006119
NM_033163
NM_033164
NM_033165
|
FGF8
|
fibroblast growth factor 8 (androgen-induced)
|
|
chr19_-_11234117
|
0.857
|
NM_020812
|
DOCK6
|
dedicator of cytokinesis 6
|
|
chr22_+_28032950
|
0.855
|
NM_006478
NM_152236
NM_152237
|
GAS2L1
|
growth arrest-specific 2 like 1
|
|
chr22_+_41877502
|
0.854
|
|
TSPO
|
translocator protein (18kDa)
|
|
chr22_+_38720881
|
0.853
|
NM_138435
|
FAM83F
|
family with sequence similarity 83, member F
|
|
chr22_+_41877470
|
0.851
|
NM_000714
NM_007311
|
TSPO
|
translocator protein (18kDa)
|
|
chr19_+_1389410
|
0.851
|
|
RPS15
|
ribosomal protein S15
|
|
chr19_+_41177882
|
0.848
|
NM_001042631
|
SDHAF1
|
succinate dehydrogenase complex assembly factor 1
|
|
chr4_-_2336328
|
0.838
|
NM_001172659
NM_001172660
|
ZFYVE28
|
zinc finger, FYVE domain containing 28
|
|
chr19_+_7874765
|
0.837
|
|
MAP2K7
|
mitogen-activated protein kinase kinase 7
|
|
chr22_-_43783449
|
0.834
|
|
PHF21B
|
PHD finger protein 21B
|
|
chr10_+_50492088
|
0.832
|
NM_001142929
NM_001142933
NM_001142934
NM_020549
|
CHAT
|
choline O-acetyltransferase
|
|
chr16_-_30274182
|
0.830
|
NM_006110
|
CD2BP2
|
CD2 (cytoplasmic tail) binding protein 2
|
|
chr2_+_119697859
|
0.828
|
|
STEAP3
|
STEAP family member 3
|
|
chr22_-_17546296
|
0.828
|
NM_005984
|
SLC25A1
|
solute carrier family 25 (mitochondrial carrier; citrate transporter), member 1
|
|
chr11_+_823007
|
0.828
|
|
CD151
|
CD151 molecule (Raph blood group)
|
|
chr16_+_517834
|
0.824
|
NM_005632
|
SOLH
|
small optic lobes homolog (Drosophila)
|
|
chr19_-_19599973
|
0.816
|
NM_004720
|
LPAR2
|
lysophosphatidic acid receptor 2
|
|
chr19_+_50963484
|
0.815
|
|
|
|
|
chr12_+_130945215
|
0.815
|
NM_003565
|
ULK1
|
unc-51-like kinase 1 (C. elegans)
|
|
chr19_+_7874774
|
0.813
|
|
MAP2K7
|
mitogen-activated protein kinase kinase 7
|
|
chr16_-_342399
|
0.812
|
NM_003502
NM_181050
|
AXIN1
|
axin 1
|
|
chr19_+_3310138
|
0.808
|
NM_205843
|
NFIC
|
nuclear factor I/C (CCAAT-binding transcription factor)
|
|
chr19_-_41397396
|
0.805
|
NM_152477
|
ZNF565
|
zinc finger protein 565
|
|
chr1_-_29322993
|
0.804
|
NM_001003682
|
TMEM200B
|
transmembrane protein 200B
|
|
chr11_+_6368213
|
0.802
|
NM_000543
NM_001007593
|
SMPD1
|
sphingomyelin phosphodiesterase 1, acid lysosomal
|
|
chr2_+_238200922
|
0.801
|
NM_001137550
|
LRRFIP1
|
leucine rich repeat (in FLII) interacting protein 1
|
|
chr16_+_1299693
|
0.799
|
|
UBE2I
|
ubiquitin-conjugating enzyme E2I (UBC9 homolog, yeast)
|
|
chr9_-_139060377
|
0.797
|
|
NPDC1
|
neural proliferation, differentiation and control, 1
|
|
chr10_+_75340867
|
0.795
|
NM_001145031
NM_002658
|
PLAU
|
plasminogen activator, urokinase
|
|
chr6_+_30082331
|
0.795
|
|
HLA-J
|
major histocompatibility complex, class I, J (pseudogene)
|
|
chr2_+_10969513
|
0.793
|
NM_002236
|
KCNF1
|
potassium voltage-gated channel, subfamily F, member 1
|
|
chr22_-_20419975
|
0.793
|
NM_013313
|
YPEL1
|
yippee-like 1 (Drosophila)
|