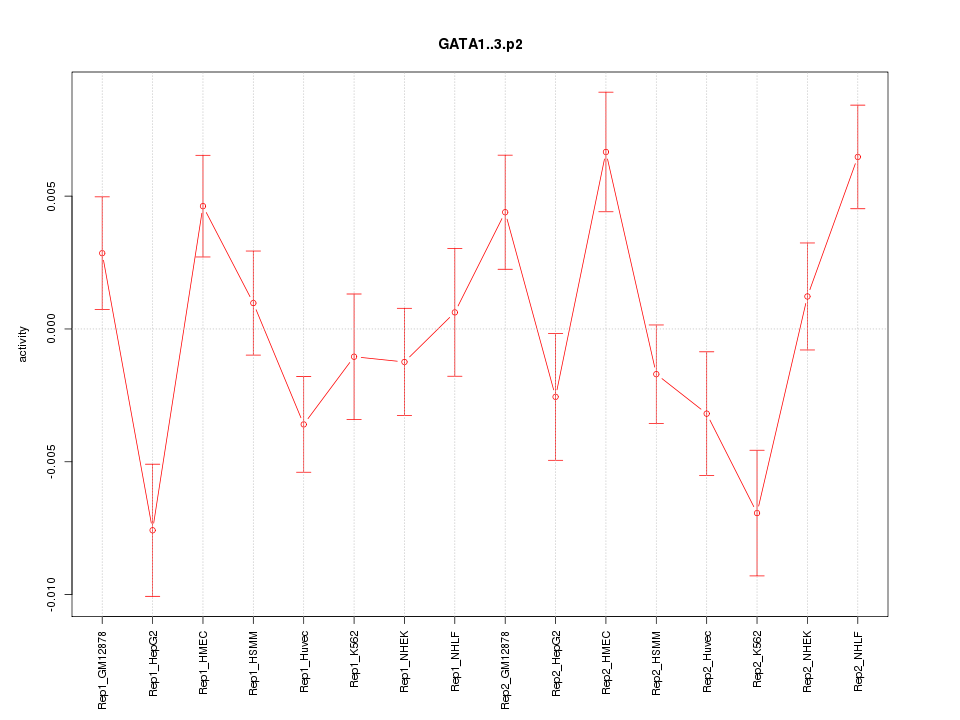
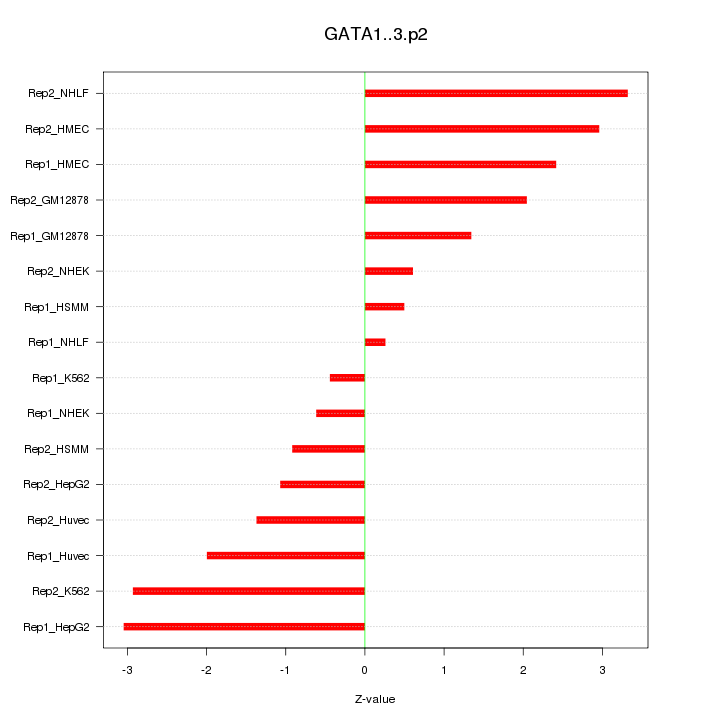
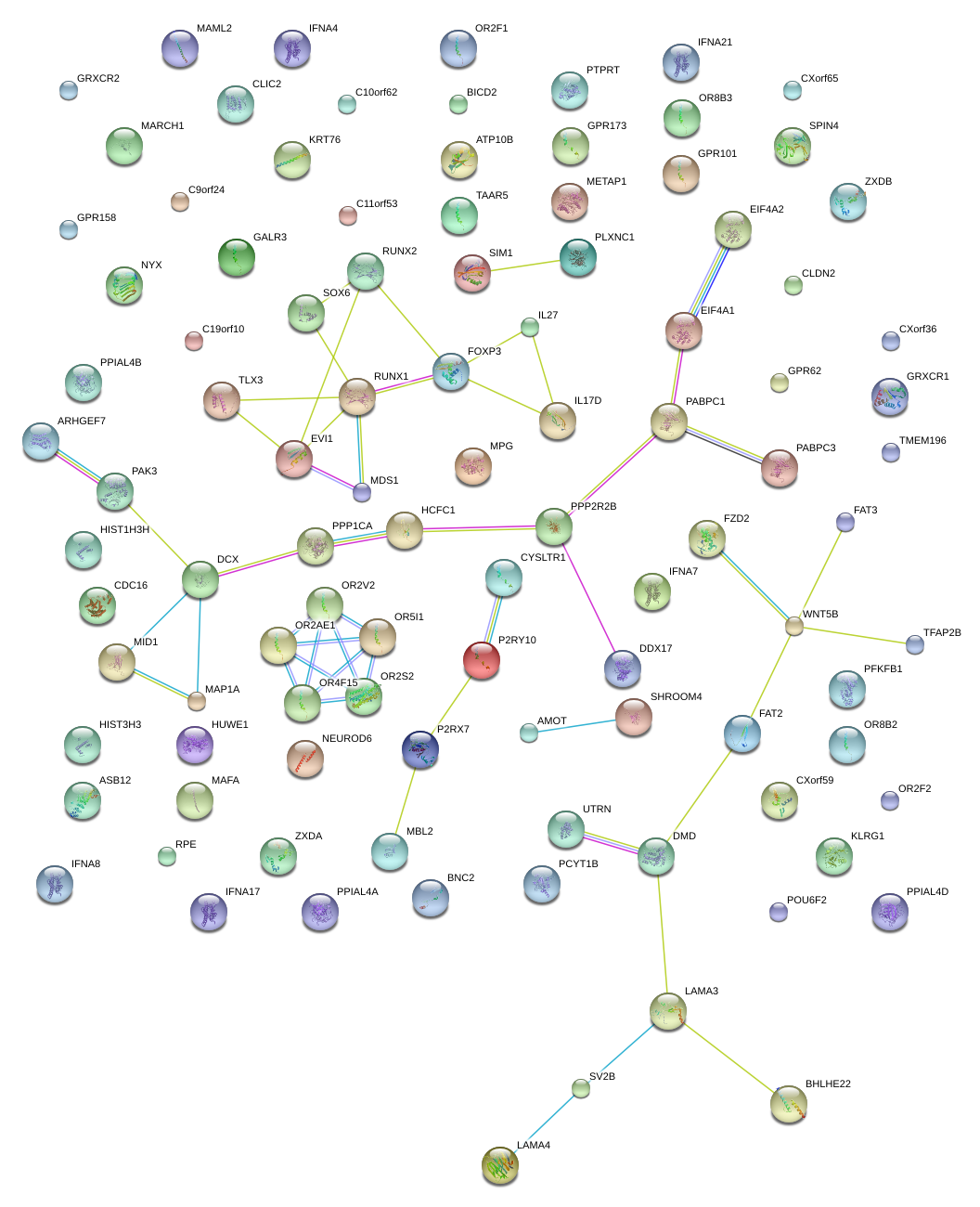
|
chr3_-_170864111
|
2.635
|
NM_004991
|
MECOM
|
MDS1 and EVI1 complex locus
|
|
chr12_+_1608672
|
2.184
|
NM_032642
|
WNT5B
|
wingless-type MMTV integration site family, member 5B
|
|
chrX_-_57953578
|
2.108
|
NM_007156
|
ZXDA
|
zinc finger, X-linked, duplicated A
|
|
chr17_-_7036490
|
2.016
|
|
|
|
|
chrX_+_53095230
|
1.980
|
NM_018969
|
GPR173
|
G protein-coupled receptor 173
|
|
chrX_-_50403412
|
1.972
|
|
SHROOM4
|
shroom family member 4
|
|
chr18_+_19706979
|
1.827
|
NM_000227
NM_001127718
|
LAMA3
|
laminin, alpha 3
|
|
chrX_-_63362227
|
1.822
|
NM_130388
|
ASB12
|
ankyrin repeat and SOCS box containing 12
|
|
chrX_+_41191630
|
1.772
|
NM_022567
|
NYX
|
nyctalopin
|
|
chr5_-_160044844
|
1.687
|
|
ATP10B
|
ATPase, class V, type 10B
|
|
chrX_-_152889815
|
1.649
|
NM_005334
|
HCFC1
|
host cell factor C1 (VP16-accessory protein)
|
|
chr15_+_100175888
|
1.581
|
NM_001001674
|
OR4F15
|
olfactory receptor, family 4, subfamily F, member 15
|
|
chrX_-_55037235
|
1.555
|
NM_002625
|
PFKFB1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1
|
|
chr21_-_35153664
|
1.535
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chrX_+_110252960
|
1.532
|
NM_001128168
NM_001128172
NM_001128173
|
PAK3
|
p21 protein (Cdc42/Rac)-activated kinase 3
|
|
chr12_+_93066370
|
1.531
|
NM_005761
|
PLXNC1
|
plexin C1
|
|
chr5_+_180514548
|
1.522
|
NM_206880
|
OR2V2
|
olfactory receptor, family 2, subfamily V, member 2
|
|
chrX_-_110541029
|
1.450
|
NM_000555
|
DCX
|
doublecortin
|
|
chr9_-_34387786
|
1.438
|
NM_032596
|
C9orf24
|
chromosome 9 open reading frame 24
|
|
chr11_-_16380967
|
1.427
|
NM_001145819
NM_017508
|
SOX6
|
SRY (sex determining region Y)-box 6
|
|
chr1_-_226679648
|
1.359
|
NM_003493
|
HIST3H3
|
histone cluster 3, H3
|
|
chr5_+_170668892
|
1.352
|
NM_021025
|
TLX3
|
T-cell leukemia homeobox 3
|
|
chr17_+_39990124
|
1.316
|
NM_001466
|
FZD2
|
frizzled homolog 2 (Drosophila)
|
|
chr10_-_54201400
|
1.311
|
NM_000242
|
MBL2
|
mannose-binding lectin (protein C) 2, soluble
|
|
chr11_+_59001421
|
1.253
|
NM_001004705
|
OR4D10
|
olfactory receptor, family 4, subfamily D, member 10
|
|
chr7_+_38984133
|
1.227
|
NM_001166018
NM_007252
|
POU6F2
|
POU class 6 homeobox 2
|
|
chrX_-_44945025
|
1.205
|
NM_024689
NM_176819
|
CXorf36
|
chromosome X open reading frame 36
|
|
chrX_+_110074124
|
1.186
|
NM_001128166
NM_001128167
|
PAK3
|
p21 protein (Cdc42/Rac)-activated kinase 3
|
|
chr9_+_21399132
|
1.177
|
NM_002170
|
IFNA8
|
interferon, alpha 8
|
|
chrX_-_39532391
|
1.166
|
|
|
|
|
chrX_+_106057984
|
1.148
|
|
CLDN2
|
claudin 2
|
|
chr9_-_16717887
|
1.098
|
|
BNC2
|
basonuclin 2
|
|
chr5_-_145232723
|
1.097
|
NM_001080516
|
GRXCR2
|
glutaredoxin, cysteine rich 2
|
|
chr11_+_110631916
|
1.095
|
NM_198498
|
C11orf53
|
chromosome 11 open reading frame 53
|
|
chr9_-_35948150
|
1.095
|
NM_019897
|
OR2S2
|
olfactory receptor, family 2, subfamily S, member 2
|
|
chr22_-_37211467
|
1.085
|
|
DDX17
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 17
|
|
chrX_-_24600661
|
1.082
|
NM_001163264
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta
|
|
chr16_-_28425632
|
1.070
|
NM_145659
|
IL27
|
interleukin 27
|
|
chrX_-_70243362
|
1.061
|
NM_001025265
|
CXorf65
|
chromosome X open reading frame 65
|
|
chr6_-_132952517
|
1.049
|
NM_003967
|
TAAR5
|
trace amine associated receptor 5
|
|
chr4_+_42590038
|
1.049
|
NM_001080476
|
GRXCR1
|
glutaredoxin, cysteine rich 1
|
|
chrX_-_53646332
|
1.046
|
|
HUWE1
|
HECT, UBA and WWE domain containing 1
|
|
chr9_-_94566823
|
1.044
|
NM_001003800
NM_015250
|
BICD2
|
bicaudal D homolog 2 (Drosophila)
|
|
chr7_-_31347032
|
1.037
|
NM_022728
|
NEUROD6
|
neurogenic differentiation 6
|
|
chr7_+_143263191
|
1.017
|
NM_001004685
|
OR2F2
|
olfactory receptor, family 2, subfamily F, member 2
|
|
chr10_+_104995633
|
1.010
|
NM_001143909
|
LOC729020
|
rcRPE
|
|
chrX_+_35974973
|
1.005
|
NM_173695
|
CXorf59
|
chromosome X open reading frame 59
|
|
chr11_-_95715976
|
0.997
|
NM_032427
|
MAML2
|
mastermind-like 2 (Drosophila)
|
|
chrX_-_33267646
|
0.996
|
NM_000109
|
DMD
|
dystrophin
|
|
chr7_-_19778928
|
0.992
|
NM_152774
|
TMEM196
|
transmembrane protein 196
|
|
chr12_-_51457371
|
0.981
|
NM_015848
|
KRT76
|
keratin 76
|
|
chr14_-_100420936
|
0.974
|
NM_001134888
|
RTL1
|
retrotransposon-like 1
|
|
chr8_+_65655367
|
0.969
|
NM_152414
|
BHLHE22
|
basic helix-loop-helix family, member e22
|
|
chr9_-_21177529
|
0.968
|
NM_021068
|
IFNA4
|
interferon, alpha 4
|
|
chrX_-_49008115
|
0.957
|
NM_001114377
NM_014009
|
FOXP3
|
forkhead box P3
|
|
chrX_-_111970662
|
0.953
|
NM_133265
|
AMOT
|
angiomotin
|
|
chr6_-_101018271
|
0.936
|
NM_005068
|
SIM1
|
single-minded homolog 1 (Drosophila)
|
|
chrX_-_10605726
|
0.933
|
NM_001098624
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chr5_-_146415782
|
0.931
|
NM_181674
NM_181676
|
PPP2R2B
|
protein phosphatase 2, regulatory subunit B, beta
|
|
chrX_+_78087484
|
0.910
|
NM_014499
NM_198333
|
P2RY10
|
purinergic receptor P2Y, G-protein coupled, 10
|
|
chr6_+_50894397
|
0.910
|
NM_003221
|
TFAP2B
|
transcription factor AP-2 beta (activating enhancer binding protein 2 beta)
|
|
chr10_+_99339344
|
0.909
|
NM_001009997
|
C10orf62
|
chromosome 10 open reading frame 62
|
|
chr7_-_99312591
|
0.902
|
NM_001005276
|
OR2AE1
|
olfactory receptor, family 2, subfamily AE, member 1
|
|
chr13_+_24568275
|
0.887
|
NM_030979
|
PABPC3
|
poly(A) binding protein, cytoplasmic 3
|
|
chr1_-_146569159
|
0.883
|
NM_001164261
NM_001164262
NM_001144032
|
PPIAL4D
PPIAL4F
PPIAL4E
|
peptidylprolyl isomerase A (cyclophilin A)-like 4D
peptidylprolyl isomerase A (cyclophilin A)-like 4F
peptidylprolyl isomerase A (cyclophilin A)-like 4E
|
|
chr8_-_144583718
|
0.883
|
NM_201589
|
MAFA
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog A (avian)
|
|
chrX_-_111953009
|
0.877
|
NM_001113490
|
AMOT
|
angiomotin
|
|
chrX_-_135941498
|
0.871
|
NM_054021
|
GPR101
|
G protein-coupled receptor 101
|
|
chr1_+_109450116
|
0.858
|
|
|
|
|
chrX_-_62487693
|
0.846
|
NM_001012968
|
SPIN4
|
spindlin family, member 4
|
|
chr15_+_89570333
|
0.842
|
|
SV2B
|
synaptic vesicle glycoprotein 2B
|
|
chr11_-_123772456
|
0.839
|
NM_001005467
|
OR8B3
|
olfactory receptor, family 8, subfamily B, member 3
|
|
chr10_+_25504295
|
0.837
|
NM_020752
|
GPR158
|
G protein-coupled receptor 158
|
|
chr15_+_41597097
|
0.836
|
NM_002373
|
MAP1A
|
microtubule-associated protein 1A
|
|
chrX_-_10495476
|
0.834
|
NM_001193278
NM_001193279
NM_001193280
NM_001193281
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chr3_+_51964369
|
0.832
|
NM_080865
|
GPR62
|
G protein-coupled receptor 62
|
|
chrX_-_154216929
|
0.830
|
NM_001289
|
CLIC2
|
chloride intracellular channel 2
|
|
chr4_-_164754106
|
0.828
|
NM_017923
|
MARCH1
|
membrane-associated ring finger (C3HC4) 1
|
|
chr11_+_91724909
|
0.824
|
NM_001008781
|
FAT3
|
FAT tumor suppressor homolog 3 (Drosophila)
|
|
chr22_+_36549334
|
0.819
|
NM_003614
|
GALR3
|
galanin receptor 3
|
|
chr17_+_7416780
|
0.817
|
NM_001416
|
EIF4A1
|
eukaryotic translation initiation factor 4A1
|
|
chrX_-_50573759
|
0.808
|
NM_020717
|
SHROOM4
|
shroom family member 4
|
|
chrX_-_77469635
|
0.805
|
NM_006639
|
CYSLTR1
|
cysteinyl leukotriene receptor 1
|
|
chr11_-_55460451
|
0.803
|
NM_006637
|
OR5I1
|
olfactory receptor, family 5, subfamily I, member 1
|
|
chr14_+_59934178
|
0.802
|
|
|
|
|
chr11_+_55776251
|
0.800
|
NM_001004747
|
OR5T3
|
olfactory receptor, family 5, subfamily T, member 3
|
|
chr17_-_3248453
|
0.797
|
NM_003553
|
OR1E1
|
olfactory receptor, family 1, subfamily E, member 1
|
|
chrX_+_102498035
|
0.797
|
NM_001006612
NM_001006613
NM_001006614
NM_016303
|
WBP5
|
WW domain binding protein 5
|
|
chr11_-_19038803
|
0.792
|
NM_054030
|
MRGPRX2
|
MAS-related GPR, member X2
|
|
chr4_-_41445479
|
0.791
|
|
PHOX2B
|
paired-like homeobox 2b
|
|
chr11_-_57643491
|
0.784
|
NM_001005211
|
OR9I1
|
olfactory receptor, family 9, subfamily I, member 1
|
|
chr14_-_60260211
|
0.782
|
NM_017420
|
SIX4
|
SIX homeobox 4
|
|
chrX_-_71850434
|
0.776
|
NM_001122670
NM_001172436
NM_002637
|
PHKA1
|
phosphorylase kinase, alpha 1 (muscle)
|
|
chr9_-_21207303
|
0.775
|
NM_002173
|
IFNA16
|
interferon, alpha 16
|
|
chr15_+_87540238
|
0.771
|
|
ABHD2
|
abhydrolase domain containing 2
|
|
chr13_-_45640679
|
0.769
|
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin)
|
|
chr12_+_79634820
|
0.769
|
NM_005593
|
MYF5
|
myogenic factor 5
|
|
chr15_+_100163424
|
0.765
|
NM_001005326
|
OR4F6
|
olfactory receptor, family 4, subfamily F, member 6
|
|
chrX_+_135397790
|
0.758
|
NM_001727
|
BRS3
|
bombesin-like receptor 3
|
|
chr10_-_21847126
|
0.754
|
|
|
|
|
chrX_+_129133234
|
0.754
|
NM_004794
|
RAB33A
|
RAB33A, member RAS oncogene family
|
|
chrX_-_138552517
|
0.750
|
NM_001171877
NM_001171878
NM_001171879
NM_005369
|
MCF2
|
MCF.2 cell line derived transforming sequence
|
|
chrX_-_21686079
|
0.744
|
NM_014332
|
SMPX
|
small muscle protein, X-linked
|
|
chr11_-_6148131
|
0.740
|
NM_001004052
|
OR52B2
|
olfactory receptor, family 52, subfamily B, member 2
|
|
chr12_-_98072602
|
0.739
|
NM_020140
NM_181670
|
ANKS1B
|
ankyrin repeat and sterile alpha motif domain containing 1B
|
|
chr17_-_9635325
|
0.738
|
NM_001105571
|
DHRS7C
|
dehydrogenase/reductase (SDR family) member 7C
|
|
chrX_+_49013424
|
0.738
|
|
PPP1R3F
|
protein phosphatase 1, regulatory (inhibitor) subunit 3F
|
|
chrX_+_48253007
|
0.734
|
NM_203474
NM_203475
|
PORCN
|
porcupine homolog (Drosophila)
|
|
chrX_+_105823723
|
0.724
|
NM_024539
|
RNF128
|
ring finger protein 128
|
|
chrX_+_57634993
|
0.722
|
NM_007157
|
ZXDB
|
zinc finger, X-linked, duplicated B
|
|
chr2_-_39040981
|
0.721
|
|
LOC375196
|
hypothetical LOC375196
|
|
chr10_-_119124727
|
0.718
|
NM_173791
|
PDZD8
|
PDZ domain containing 8
|
|
chr6_+_108593907
|
0.707
|
NM_003269
|
NR2E1
|
nuclear receptor subfamily 2, group E, member 1
|
|
chr8_+_62363078
|
0.705
|
NM_173519
|
CLVS1
|
clavesin 1
|
|
chr11_-_10546654
|
0.702
|
NM_006691
|
LYVE1
|
lymphatic vessel endothelial hyaluronan receptor 1
|
|
chr6_-_46933901
|
0.701
|
|
GPR116
|
G protein-coupled receptor 116
|
|
chr14_-_57133361
|
0.699
|
NM_001080455
|
SLC35F4
|
solute carrier family 35, member F4
|
|
chr8_+_22266256
|
0.699
|
|
PIWIL2
|
piwi-like 2 (Drosophila)
|
|
chr11_-_8241875
|
0.692
|
NM_002315
|
LMO1
|
LIM domain only 1 (rhombotin 1)
|
|
chr19_+_54299686
|
0.690
|
|
SNRNP70
|
small nuclear ribonucleoprotein 70kDa (U1)
|
|
chr6_-_80713589
|
0.687
|
NM_022726
|
ELOVL4
|
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 4
|
|
chr11_+_59979857
|
0.683
|
NM_021950
NM_152866
|
MS4A1
|
membrane-spanning 4-domains, subfamily A, member 1
|
|
chr7_+_23496661
|
0.683
|
|
RPS2P32
|
ribosomal protein S2 pseudogene 32
|
|
chr1_+_146910634
|
0.682
|
NM_001144032
|
PPIAL4E
|
peptidylprolyl isomerase A (cyclophilin A)-like 4E
|
|
chr12_-_6995988
|
0.675
|
NM_005768
|
LPCAT3
|
lysophosphatidylcholine acyltransferase 3
|
|
chr11_+_123945102
|
0.673
|
NM_001005194
|
OR8A1
|
olfactory receptor, family 8, subfamily A, member 1
|
|
chr1_-_157003095
|
0.668
|
NM_001005185
|
OR6N1
|
olfactory receptor, family 6, subfamily N, member 1
|
|
chr12_+_48630790
|
0.667
|
NM_000486
|
AQP2
|
aquaporin 2 (collecting duct)
|
|
chrX_+_38305634
|
0.667
|
NM_004615
|
TSPAN7
|
tetraspanin 7
|
|
chrX_-_54226364
|
0.667
|
NM_017848
NM_198456
|
FAM120C
|
family with sequence similarity 120C
|
|
chr11_-_64133908
|
0.666
|
|
|
|
|
chr1_-_143075560
|
0.665
|
NM_178230
NM_001143883
NM_001135789
|
PPIAL4A
PPIAL4G
PPIAL4B
PPIAL4C
|
peptidylprolyl isomerase A (cyclophilin A)-like 4A
peptidylprolyl isomerase A (cyclophilin A)-like 4G
peptidylprolyl isomerase A (cyclophilin A)-like 4B
peptidylprolyl isomerase A (cyclophilin A)-like 4C
|
|
chr17_-_31104009
|
0.665
|
NM_139285
|
GAS2L2
|
growth arrest-specific 2 like 2
|
|
chr17_+_1120607
|
0.660
|
NM_001164405
|
BHLHA9
|
basic helix-loop-helix family, member a9
|
|
chr15_+_52092392
|
0.659
|
NM_001080534
|
UNC13C
|
unc-13 homolog C (C. elegans)
|
|
chr7_-_36992221
|
0.658
|
NM_130442
|
ELMO1
|
engulfment and cell motility 1
|
|
chr3_-_57288019
|
0.655
|
NM_130387
|
ASB14
|
ankyrin repeat and SOCS box containing 14
|
|
chr9_-_34371497
|
0.652
|
NM_147168
NM_147169
|
C9orf24
|
chromosome 9 open reading frame 24
|
|
chr11_+_58967192
|
0.651
|
NM_001004728
|
OR5A1
|
olfactory receptor, family 5, subfamily A, member 1
|
|
chrX_+_66683075
|
0.651
|
|
AR
|
androgen receptor
|
|
chrX_+_69559605
|
0.649
|
NM_001171191
NM_001171192
NM_001171193
NM_017711
|
GDPD2
|
glycerophosphodiester phosphodiesterase domain containing 2
|
|
chr1_-_157551072
|
0.645
|
NM_001004467
|
OR10J3
|
olfactory receptor, family 10, subfamily J, member 3
|
|
chr18_-_55136673
|
0.644
|
NM_181654
|
CPLX4
|
complexin 4
|
|
chr11_+_55869996
|
0.643
|
NM_001002907
|
OR8K1
|
olfactory receptor, family 8, subfamily K, member 1
|
|
chr2_+_218929822
|
0.641
|
NM_198559
|
C2orf62
|
chromosome 2 open reading frame 62
|
|
chrX_+_113724806
|
0.638
|
NM_000868
|
HTR2C
|
5-hydroxytryptamine (serotonin) receptor 2C
|
|
chr11_-_57728228
|
0.638
|
NM_001004459
|
OR1S2
|
olfactory receptor, family 1, subfamily S, member 2
|
|
chrX_-_127014020
|
0.636
|
NM_138289
|
ACTRT1
|
actin-related protein T1
|
|
chr11_+_59038961
|
0.635
|
NM_001004711
|
OR4D9
|
olfactory receptor, family 4, subfamily D, member 9
|
|
chrX_+_82649924
|
0.633
|
NM_000307
|
POU3F4
|
POU class 3 homeobox 4
|
|
chr14_-_59264044
|
0.631
|
NM_206857
|
RTN1
|
reticulon 1
|
|
chrX_+_70360184
|
0.630
|
|
GJB1
|
gap junction protein, beta 1, 32kDa
|
|
chr14_-_47213906
|
0.629
|
NM_001113498
|
MDGA2
|
MAM domain containing glycosylphosphatidylinositol anchor 2
|
|
chr16_+_3345889
|
0.628
|
NM_012368
|
OR2C1
|
olfactory receptor, family 2, subfamily C, member 1
|
|
chr1_-_157014048
|
0.627
|
NM_001005278
|
OR6N2
|
olfactory receptor, family 6, subfamily N, member 2
|
|
chrX_+_150653873
|
0.627
|
NM_005140
|
CNGA2
|
cyclic nucleotide gated channel alpha 2
|
|
chr9_-_21067804
|
0.625
|
NM_002176
|
IFNB1
|
interferon, beta 1, fibroblast
|
|
chr11_+_5862076
|
0.625
|
NM_001005165
|
OR52E4
|
olfactory receptor, family 52, subfamily E, member 4
|
|
chr4_-_70539922
|
0.625
|
NM_001105677
|
UGT2A2
|
UDP glucuronosyltransferase 2 family, polypeptide A2
|
|
chr1_+_234372454
|
0.624
|
NM_003272
|
GPR137B
|
G protein-coupled receptor 137B
|
|
chr15_+_38923914
|
0.622
|
NM_001032367
|
SPINT1
|
serine peptidase inhibitor, Kunitz type 1
|
|
chrX_+_111005934
|
0.621
|
NM_001195576
NM_001195578
|
LOC100329135
|
hypothetical LOC100329135
|
|
chr10_+_115302767
|
0.621
|
NM_004132
|
HABP2
|
hyaluronan binding protein 2
|
|
chr2_+_204440753
|
0.615
|
NM_001037631
NM_005214
|
CTLA4
|
cytotoxic T-lymphocyte-associated protein 4
|
|
chr18_-_65766068
|
0.615
|
|
CD226
|
CD226 molecule
|
|
chr1_-_26553168
|
0.612
|
NM_001039775
|
AIM1L
|
absent in melanoma 1-like
|
|
chr13_+_27264779
|
0.611
|
NM_145657
|
GSX1
|
GS homeobox 1
|
|
chr19_-_43926953
|
0.609
|
NM_144691
|
CAPN12
|
calpain 12
|
|
chrX_+_57635165
|
0.608
|
|
ZXDB
|
zinc finger, X-linked, duplicated B
|
|
chr1_+_158426908
|
0.604
|
NM_001231
|
CASQ1
|
calsequestrin 1 (fast-twitch, skeletal muscle)
|
|
chr8_-_42817604
|
0.603
|
NM_018105
NM_199003
|
THAP1
|
THAP domain containing, apoptosis associated protein 1
|
|
chr17_-_75427644
|
0.601
|
NM_003655
|
CBX4
|
chromobox homolog 4
|
|
chrX_+_70232362
|
0.597
|
NM_001170931
NM_005938
|
FOXO4
|
forkhead box O4
|
|
chr14_+_54291255
|
0.594
|
NM_001161577
|
SAMD4A
|
sterile alpha motif domain containing 4A
|
|
chr5_-_151764857
|
0.592
|
NM_020167
|
NMUR2
|
neuromedin U receptor 2
|
|
chrX_-_100527799
|
0.591
|
NM_000061
|
BTK
|
Bruton agammaglobulinemia tyrosine kinase
|
|
chr17_-_77824849
|
0.590
|
NM_001893
NM_139062
|
CSNK1D
|
casein kinase 1, delta
|
|
chrX_+_122145776
|
0.590
|
NM_000828
NM_007325
|
GRIA3
|
glutamate receptor, ionotrophic, AMPA 3
|
|
chr5_-_149649380
|
0.590
|
|
CAMK2A
|
calcium/calmodulin-dependent protein kinase II alpha
|
|
chrX_+_70438352
|
0.587
|
NM_012278
|
ITGB1BP2
|
integrin beta 1 binding protein (melusin) 2
|
|
chrX_+_51503219
|
0.584
|
NM_018094
|
GSPT2
|
G1 to S phase transition 2
|
|
chr1_-_156816312
|
0.583
|
NM_001004477
|
OR10X1
|
olfactory receptor, family 10, subfamily X, member 1
|
|
chr9_+_27099138
|
0.581
|
NM_000459
|
TEK
|
TEK tyrosine kinase, endothelial
|
|
chr3_-_49868889
|
0.578
|
NM_005879
|
TRAIP
|
TRAF interacting protein
|
|
chr11_-_46364681
|
0.576
|
NM_000741
|
CHRM4
|
cholinergic receptor, muscarinic 4
|
|
chr1_+_158317967
|
0.573
|
NM_004983
|
KCNJ9
|
potassium inwardly-rectifying channel, subfamily J, member 9
|
|
chr9_-_21340885
|
0.572
|
NM_021002
|
IFNA6
|
interferon, alpha 6
|
|
chr9_+_36159384
|
0.571
|
NM_005893
|
CCIN
|
calicin
|
|
chr7_-_81237162
|
0.571
|
NM_000601
NM_001010931
NM_001010932
NM_001010933
NM_001010934
|
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor)
|
|
chr3_-_52065130
|
0.571
|
NM_001947
|
DUSP7
|
dual specificity phosphatase 7
|
|
chrX_-_131401319
|
0.570
|
NM_018388
NM_133486
|
MBNL3
|
muscleblind-like 3 (Drosophila)
|
|
chr12_-_85756766
|
0.570
|
NM_013244
|
MGAT4C
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative)
|
|
chr5_-_146441184
|
0.568
|
NM_181677
NM_181678
|
PPP2R2B
|
protein phosphatase 2, regulatory subunit B, beta
|
|
chr12_+_5473558
|
0.568
|
NM_002527
|
NTF3
|
neurotrophin 3
|
|
chr11_+_130745580
|
0.566
|
NM_001048209
|
NTM
|
neurotrimin
|
|
chr4_-_123761615
|
0.565
|
NM_021803
|
IL21
|
interleukin 21
|
|
chr12_-_46883341
|
0.560
|
NM_001004134
|
OR10AD1
|
olfactory receptor, family 10, subfamily AD, member 1
|
|
chrX_+_49013270
|
0.558
|
|
PPP1R3F
|
protein phosphatase 1, regulatory (inhibitor) subunit 3F
|
|
chrX_+_123307812
|
0.556
|
NM_001114937
NM_002351
|
SH2D1A
|
SH2 domain containing 1A
|
|
chr4_+_76700281
|
0.554
|
NM_178497
|
C4orf26
|
chromosome 4 open reading frame 26
|