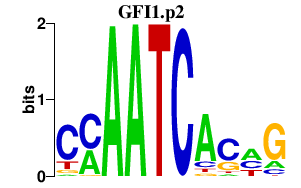
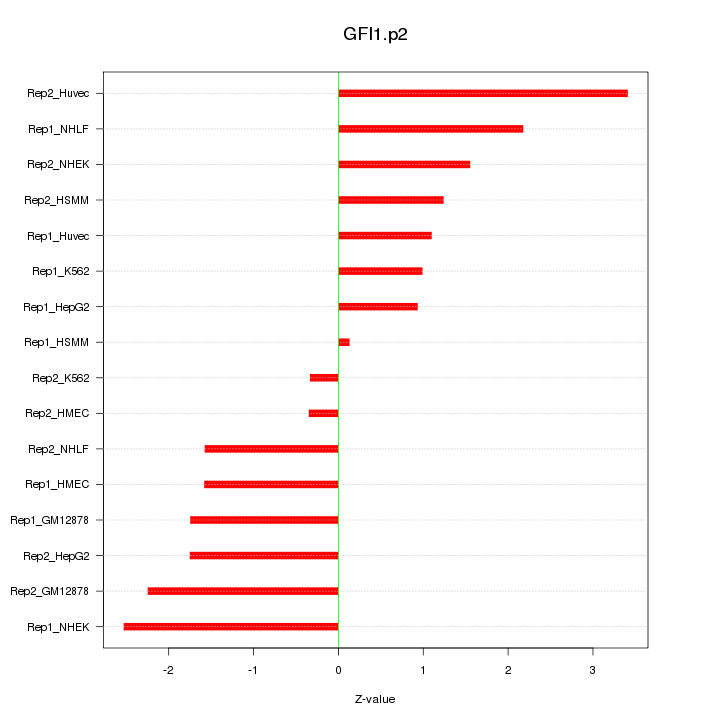
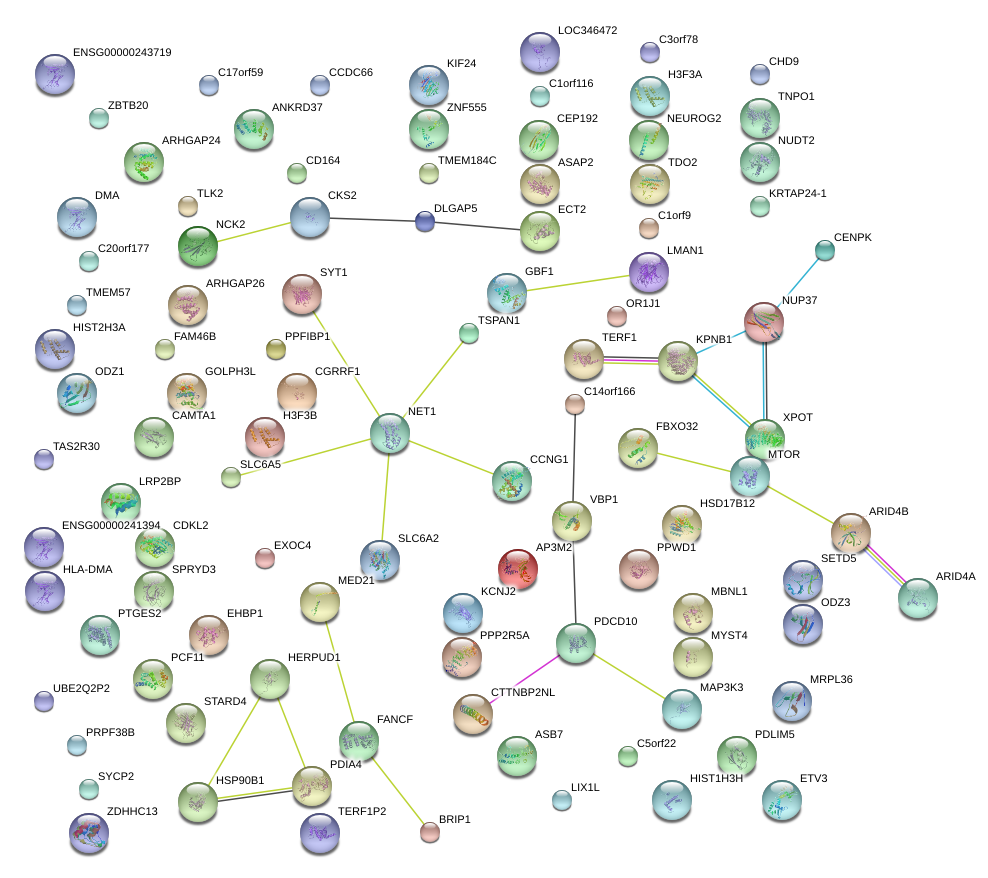
|
chr8_+_42129620
|
3.453
|
NM_001134296
NM_006803
|
AP3M2
|
adaptor-related protein complex 3, mu 2 subunit
|
|
chr17_+_59053518
|
2.162
|
NM_002401
NM_203351
|
MAP3K3
|
mitogen-activated protein kinase kinase kinase 3
|
|
chr17_+_59053515
|
2.033
|
|
MAP3K3
|
mitogen-activated protein kinase kinase kinase 3
|
|
chr17_+_59053447
|
2.033
|
|
MAP3K3
|
mitogen-activated protein kinase kinase kinase 3
|
|
chr3_+_173951163
|
1.556
|
|
ECT2
|
epithelial cell transforming sequence 2 oncogene
|
|
chr4_+_183302105
|
1.541
|
|
ODZ3
|
odz, odd Oz/ten-m homolog 3 (Drosophila)
|
|
chr3_+_153499883
|
1.534
|
NM_207293
NM_207294
NM_207295
NM_207296
NM_207297
|
MBNL1
|
muscleblind-like (Drosophila)
|
|
chr17_-_71287417
|
1.477
|
NM_005324
|
H3F3B
|
H3 histone, family 3B (H3.3B)
|
|
chr5_+_162797149
|
1.251
|
NM_004060
NM_199246
|
CCNG1
|
cyclin G1
|
|
chr10_+_5444516
|
1.247
|
NM_001047160
|
NET1
|
neuroepithelial cell transforming 1
|
|
chr17_+_43082273
|
1.242
|
NM_002265
|
KPNB1
|
karyopherin (importin) beta 1
|
|
chr3_-_116348780
|
1.203
|
NM_015642
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr14_+_57835028
|
1.180
|
|
ARID4A
|
AT rich interactive domain 4A (RBP1-like)
|
|
chr8_+_74083615
|
1.170
|
NM_003218
NM_017489
|
TERF1
|
telomeric repeat binding factor (NIMA-interacting) 1
|
|
chr14_+_57834962
|
1.139
|
NM_002892
NM_023000
NM_023001
|
ARID4A
|
AT rich interactive domain 4A (RBP1-like)
|
|
chr17_+_43082302
|
1.138
|
|
KPNB1
|
karyopherin (importin) beta 1
|
|
chr1_-_205272619
|
1.137
|
NM_001083924
NM_023938
|
C1orf116
|
chromosome 1 open reading frame 116
|
|
chr14_-_54727958
|
1.131
|
NM_001146015
NM_014750
|
DLGAP5
|
discs, large (Drosophila) homolog-associated protein 5
|
|
chr1_+_112740378
|
1.117
|
|
|
|
|
chr1_+_25629968
|
1.103
|
NM_018202
|
TMEM57
|
transmembrane protein 57
|
|
chr4_+_86968069
|
1.091
|
|
ARHGAP24
|
Rho GTPase activating protein 24
|
|
chr1_+_6767968
|
1.075
|
NM_001195563
NM_015215
|
CAMTA1
|
calmodulin binding transcription activator 1
|
|
chr17_-_8034071
|
1.034
|
|
C17orf59
|
chromosome 17 open reading frame 59
|
|
chr17_-_8034217
|
1.026
|
NM_017622
|
C17orf59
|
chromosome 17 open reading frame 59
|
|
chr17_-_57295451
|
1.020
|
NM_032043
|
BRIP1
|
BRCA1 interacting protein C-terminal helicase 1
|
|
chr1_+_112740322
|
1.019
|
NM_018704
|
CTTNBP2NL
|
CTTNBP2 N-terminal like
|
|
chr16_+_51722285
|
1.015
|
|
CHD9
|
chromodomain helicase DNA binding protein 9
|
|
chr1_+_112740380
|
1.007
|
|
LOC100129406
CTTNBP2NL
|
hypothetical protein LOC100129406
CTTNBP2 N-terminal like
|
|
chr3_+_121296290
|
1.001
|
|
|
|
|
chr5_-_64894687
|
0.986
|
NM_022145
|
CENPK
|
centromere protein K
|
|
chr17_+_65677255
|
0.986
|
NM_000891
|
KCNJ2
|
potassium inwardly-rectifying channel, subfamily J, member 2
|
|
chr3_+_9414290
|
0.973
|
NM_001080517
|
SETD5
|
SET domain containing 5
|
|
chr18_+_12981329
|
0.952
|
NM_032142
|
CEP192
|
centrosomal protein 192kDa
|
|
chr3_+_56566222
|
0.937
|
NM_001012506
NM_001141947
|
CCDC66
|
coiled-coil domain containing 66
|
|
chr17_+_57909979
|
0.927
|
NM_001112707
NM_006852
|
TLK2
|
tousled-like kinase 2
|
|
chr10_+_76256305
|
0.884
|
NM_012330
|
MYST4
|
MYST histone acetyltransferase (monocytic leukemia) 4
|
|
chr4_+_157044296
|
0.880
|
NM_005651
|
TDO2
|
tryptophan 2,3-dioxygenase
|
|
chr4_+_148758014
|
0.879
|
|
TMEM184C
|
transmembrane protein 184C
|
|
chr2_+_242679507
|
0.877
|
|
LOC728323
|
hypothetical LOC728323
|
|
chr5_+_64894854
|
0.871
|
NM_015342
|
PPWD1
|
peptidylprolyl isomerase domain and WD repeat containing 1
|
|
chr17_+_43081837
|
0.865
|
|
KPNB1
|
karyopherin (importin) beta 1
|
|
chr7_-_148356433
|
0.865
|
|
PDIA4
|
protein disulfide isomerase family A, member 4
|
|
chr5_+_142130475
|
0.853
|
NM_001135608
NM_015071
|
ARHGAP26
|
Rho GTPase activating protein 26
|
|
chr6_-_33028790
|
0.843
|
NM_006120
|
HLA-DMA
|
major histocompatibility complex, class II, DM alpha
|
|
chr11_+_19095215
|
0.835
|
NM_001001483
NM_019028
|
ZDHHC13
|
zinc finger, DHHC-type containing 13
|
|
chr17_+_43082557
|
0.827
|
|
KPNB1
|
karyopherin (importin) beta 1
|
|
chr2_+_62786290
|
0.812
|
NM_001142616
NM_015252
|
EHBP1
|
EH domain binding protein 1
|
|
chr1_-_148936123
|
0.810
|
NM_018178
|
GOLPH3L
|
golgi phosphoprotein 3-like
|
|
chr9_-_34319151
|
0.808
|
NM_194313
|
KIF24
|
kinesin family member 24
|
|
chr20_+_57942213
|
0.807
|
NM_001190826
|
C20orf177
|
chromosome 20 open reading frame 177
|
|
chr1_+_144188375
|
0.805
|
NM_153713
|
LIX1L
|
Lix1 homolog (mouse)-like
|
|
chr20_-_57942063
|
0.804
|
|
SYCP2
|
synaptonemal complex protein 2
|
|
chr4_+_148757958
|
0.797
|
NM_018241
|
TMEM184C
|
transmembrane protein 184C
|
|
chr3_-_168935287
|
0.788
|
NM_007217
NM_145860
NM_145859
|
PDCD10
|
programmed cell death 10
|
|
chr11_+_43658718
|
0.783
|
NM_016142
|
HSD17B12
|
hydroxysteroid (17-beta) dehydrogenase 12
|
|
chr17_+_20712337
|
0.776
|
|
|
|
|
chr21_-_30577120
|
0.774
|
NM_001085455
|
KRTAP24-1
|
keratin associated protein 24-1
|
|
chr2_+_105834635
|
0.770
|
NM_001004720
|
NCK2
|
NCK adaptor protein 2
|
|
chr3_-_9414122
|
0.768
|
|
LOC440944
|
hypothetical LOC440944
|
|
chr15_+_98960277
|
0.767
|
NM_024708
NM_198243
|
ASB7
|
ankyrin repeat and SOCS box containing 7
|
|
chr7_-_148356697
|
0.763
|
NM_004911
|
PDIA4
|
protein disulfide isomerase family A, member 4
|
|
chr5_+_31568129
|
0.762
|
NM_018356
|
C5orf22
|
chromosome 5 open reading frame 22
|
|
chr12_+_27066768
|
0.758
|
|
MED21
|
mediator complex subunit 21
|
|
chr9_+_91115924
|
0.748
|
NM_001827
|
CKS2
|
CDC28 protein kinase regulatory subunit 2
|
|
chr12_+_63090008
|
0.747
|
|
XPOT
|
exportin, tRNA (nuclear export receptor for tRNAs)
|
|
chr6_-_109795925
|
0.744
|
|
CD164
|
CD164 molecule, sialomucin
|
|
chr12_+_102848344
|
0.743
|
|
HSP90B1
|
heat shock protein 90kDa beta (Grp94), member 1
|
|
chr11_+_82545784
|
0.733
|
NM_015885
|
PCF11
|
PCF11, cleavage and polyadenylation factor subunit, homolog (S. cerevisiae)
|
|
chr4_-_113656762
|
0.723
|
NM_024019
|
NEUROG2
|
neurogenin 2
|
|
chr12_+_102848316
|
0.721
|
NM_003299
|
HSP90B1
|
heat shock protein 90kDa beta (Grp94), member 1
|
|
chr7_-_148356664
|
0.719
|
|
PDIA4
|
protein disulfide isomerase family A, member 4
|
|
chr12_+_27568311
|
0.718
|
NM_001198915
NM_001198916
NM_003622
NM_177444
|
PPFIBP1
|
PTPRF interacting protein, binding protein 1 (liprin beta 1)
|
|
chr9_+_34319503
|
0.718
|
NM_001161
NM_147172
NM_147173
|
NUDT2
|
nudix (nucleoside diphosphate linked moiety X)-type motif 2
|
|
chr14_+_51526071
|
0.717
|
|
C14orf166
|
chromosome 14 open reading frame 166
|
|
chr4_-_76774594
|
0.716
|
NM_003948
|
CDKL2
|
cyclin-dependent kinase-like 2 (CDC2-related kinase)
|
|
chr5_+_72148173
|
0.715
|
NM_002270
|
TNPO1
|
transportin 1
|
|
chr14_+_54046295
|
0.714
|
NM_006568
|
CGRRF1
|
cell growth regulator with ring finger domain 1
|
|
chr12_+_102848322
|
0.707
|
|
HSP90B1
|
heat shock protein 90kDa beta (Grp94), member 1
|
|
chrX_+_154097894
|
0.707
|
NM_003372
|
VBP1
|
von Hippel-Lindau binding protein 1
|
|
chr16_+_55523526
|
0.707
|
|
HERPUD1
|
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1
|
|
chr14_+_51525942
|
0.707
|
NM_016039
|
C14orf166
|
chromosome 14 open reading frame 166
|
|
chr4_-_186537145
|
0.706
|
NM_018409
|
LRP2BP
|
LRP2 binding protein
|
|
chr5_+_142130154
|
0.696
|
|
ARHGAP26
|
Rho GTPase activating protein 26
|
|
chr1_+_170768914
|
0.694
|
NM_016227
|
C1orf9
|
chromosome 1 open reading frame 9
|
|
chr12_-_101037976
|
0.690
|
|
NUP37
|
nucleoporin 37kDa
|
|
chr10_+_103995297
|
0.687
|
NM_004193
|
GBF1
|
golgi brefeldin A resistant guanine nucleotide exchange factor 1
|
|
chr1_-_27212035
|
0.687
|
|
FAM46B
|
family with sequence similarity 46, member B
|
|
chr7_+_132588359
|
0.685
|
NM_001037126
NM_021807
|
EXOC4
|
exocyst complex component 4
|
|
chr12_+_77782719
|
0.684
|
|
SYT1
|
synaptotagmin I
|
|
chr19_+_2792432
|
0.671
|
NM_001172775
NM_152791
|
ZNF555
|
zinc finger protein 555
|
|
chr1_-_11245130
|
0.668
|
|
MTOR
|
mechanistic target of rapamycin (serine/threonine kinase)
|
|
chr18_-_55177430
|
0.667
|
NM_005570
|
LMAN1
|
lectin, mannose-binding, 1
|
|
chr8_-_124613557
|
0.666
|
NM_148177
|
FBXO32
|
F-box protein 32
|
|
chr5_-_110876186
|
0.664
|
|
STARD4
|
StAR-related lipid transfer (START) domain containing 4
|
|
chr4_+_95592030
|
0.663
|
NM_001011513
NM_001011515
NM_001011516
NM_006457
|
PDLIM5
|
PDZ and LIM domain 5
|
|
chr3_+_52545660
|
0.662
|
NM_001124767
|
C3orf78
|
chromosome 3 open reading frame 78
|
|
chr2_+_9264360
|
0.660
|
|
ASAP2
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2
|
|
chr15_+_80434362
|
0.654
|
|
UBE2Q2P2
|
ubiquitin-conjugating enzyme E2Q family member 2 pseudogene 2
|
|
chr12_-_51759438
|
0.653
|
NM_032840
|
SPRYD3
|
SPRY domain containing 3
|
|
chr12_-_11075272
|
0.647
|
NM_176885
|
TAS2R31
|
taste receptor, type 2, member 31
|
|
chr1_+_210525490
|
0.646
|
NM_006243
|
PPP2R5A
|
protein phosphatase 2, regulatory subunit B', alpha
|
|
chr11_-_22603905
|
0.643
|
NM_022725
|
FANCF
|
Fanconi anemia, complementation group F
|
|
chr9_-_124280025
|
0.639
|
NM_001004451
|
OR1J1
|
olfactory receptor, family 1, subfamily J, member 1
|
|
chr10_+_103995248
|
0.636
|
|
GBF1
|
golgi brefeldin A resistant guanine nucleotide exchange factor 1
|
|
chr12_+_105873803
|
0.634
|
|
C12orf23
|
chromosome 12 open reading frame 23
|
|
chr12_-_51759434
|
0.629
|
|
SPRYD3
|
SPRY domain containing 3
|
|
chr6_-_136613097
|
0.629
|
NM_001099286
NM_138419
|
FAM54A
|
family with sequence similarity 54, member A
|
|
chr15_+_36331411
|
0.624
|
|
SPRED1
|
sprouty-related, EVH1 domain containing 1
|
|
chr5_-_40791766
|
0.624
|
NM_012382
|
TTC33
|
tetratricopeptide repeat domain 33
|
|
chr12_-_48705427
|
0.623
|
NM_001126103
NM_001126104
NM_013277
|
RACGAP1
|
Rac GTPase activating protein 1
|
|
chr3_+_112876212
|
0.621
|
NM_001185106
NM_153268
|
PLCXD2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2
|
|
chr7_+_65307816
|
0.620
|
|
TPST1
|
tyrosylprotein sulfotransferase 1
|
|
chr17_+_23394062
|
0.620
|
|
NLK
|
nemo-like kinase
|
|
chr3_+_184648089
|
0.617
|
|
LOC100505687
|
hypothetical LOC100505687
|
|
chr7_+_87095640
|
0.617
|
NM_001134405
NM_001134406
NM_138290
|
RUNDC3B
|
RUN domain containing 3B
|
|
chr5_-_157011665
|
0.615
|
|
|
|
|
chr10_-_99436889
|
0.613
|
NM_021732
|
AVPI1
|
arginine vasopressin-induced 1
|
|
chr3_+_170974745
|
0.611
|
NM_001185119
|
MYNN
|
myoneurin
|
|
chr12_+_105873669
|
0.611
|
NM_152261
|
C12orf23
|
chromosome 12 open reading frame 23
|
|
chr19_-_21826609
|
0.599
|
|
ZNF43
|
zinc finger protein 43
|
|
chr7_+_106596641
|
0.596
|
NM_012257
|
HBP1
|
HMG-box transcription factor 1
|
|
chr11_-_5037432
|
0.593
|
NM_001005164
|
OR52E2
|
olfactory receptor, family 52, subfamily E, member 2
|
|
chr12_+_102848287
|
0.593
|
|
HSP90B1
|
heat shock protein 90kDa beta (Grp94), member 1
|
|
chr11_+_82290384
|
0.592
|
NM_145018
|
C11orf82
|
chromosome 11 open reading frame 82
|
|
chr6_+_99389300
|
0.587
|
NM_005604
|
POU3F2
|
POU class 3 homeobox 2
|
|
chr13_+_99539269
|
0.586
|
NM_000282
NM_001127692
NM_001178004
|
PCCA
|
propionyl CoA carboxylase, alpha polypeptide
|
|
chr17_+_64019693
|
0.583
|
NM_212471
|
PRKAR1A
|
protein kinase, cAMP-dependent, regulatory, type I, alpha (tissue specific extinguisher 1)
|
|
chr7_-_86686754
|
0.582
|
NM_024315
|
C7orf23
|
chromosome 7 open reading frame 23
|
|
chr3_+_56566277
|
0.581
|
|
CCDC66
|
coiled-coil domain containing 66
|
|
chr12_+_94776836
|
0.580
|
NM_003095
|
SNRPF
|
small nuclear ribonucleoprotein polypeptide F
|
|
chr7_+_99974781
|
0.577
|
|
AGFG2
|
ArfGAP with FG repeats 2
|
|
chr21_+_43186428
|
0.576
|
NM_001001503
NM_021075
|
NDUFV3
|
NADH dehydrogenase (ubiquinone) flavoprotein 3, 10kDa
|
|
chr12_+_48303607
|
0.574
|
NM_012272
|
PRPF40B
|
PRP40 pre-mRNA processing factor 40 homolog B (S. cerevisiae)
|
|
chr11_+_31487872
|
0.572
|
NM_019040
|
ELP4
|
elongation protein 4 homolog (S. cerevisiae)
|
|
chr4_+_95592067
|
0.571
|
|
PDLIM5
|
PDZ and LIM domain 5
|
|
chr17_+_43540124
|
0.571
|
|
SNX11
|
sorting nexin 11
|
|
chr12_-_11106159
|
0.569
|
NM_176887
|
TAS2R46
|
taste receptor, type 2, member 46
|
|
chr1_-_11245172
|
0.568
|
|
MTOR
|
mechanistic target of rapamycin (serine/threonine kinase)
|
|
chr7_+_65307691
|
0.567
|
NM_003596
|
TPST1
|
tyrosylprotein sulfotransferase 1
|
|
chr5_-_39238818
|
0.563
|
|
FYB
|
FYN binding protein
|
|
chr8_+_67849972
|
0.561
|
NM_013257
NM_170709
|
SGK3
|
serum/glucocorticoid regulated kinase family, member 3
|
|
chr5_-_110876055
|
0.560
|
NM_139164
|
STARD4
|
StAR-related lipid transfer (START) domain containing 4
|
|
chr3_-_187771025
|
0.555
|
NM_001134415
|
TBCCD1
|
TBCC domain containing 1
|
|
chr7_-_104440707
|
0.553
|
|
LOC100216545
|
hypothetical LOC100216545
|
|
chr1_-_27211913
|
0.553
|
NM_052943
|
FAM46B
|
family with sequence similarity 46, member B
|
|
chr10_+_120779217
|
0.552
|
NM_199461
|
NANOS1
|
nanos homolog 1 (Drosophila)
|
|
chr1_+_179324458
|
0.551
|
|
IER5
|
immediate early response 5
|
|
chr5_+_60663842
|
0.550
|
NM_020928
|
ZSWIM6
|
zinc finger, SWIM-type containing 6
|
|
chr4_+_177477997
|
0.550
|
NM_021928
|
SPCS3
|
signal peptidase complex subunit 3 homolog (S. cerevisiae)
|
|
chr15_+_39310757
|
0.549
|
|
CHP
|
calcium binding protein P22
|
|
chr17_+_70964258
|
0.547
|
NM_014738
|
KIAA0195
|
KIAA0195
|
|
chr11_+_7463175
|
0.546
|
NM_198474
|
OLFML1
|
olfactomedin-like 1
|
|
chr14_+_101346032
|
0.546
|
|
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma
|
|
chr3_-_27473248
|
0.545
|
NM_003615
|
SLC4A7
|
solute carrier family 4, sodium bicarbonate cotransporter, member 7
|
|
chr4_+_177478103
|
0.544
|
|
SPCS3
|
signal peptidase complex subunit 3 homolog (S. cerevisiae)
|
|
chr10_+_112669349
|
0.543
|
|
SHOC2
|
soc-2 suppressor of clear homolog (C. elegans)
|
|
chr3_+_142587973
|
0.543
|
|
|
|
|
chr7_+_91996022
|
0.543
|
NM_032120
|
C7orf64
|
chromosome 7 open reading frame 64
|
|
chrX_+_133334965
|
0.542
|
NM_001015877
NM_032335
NM_032458
|
PHF6
|
PHD finger protein 6
|
|
chr9_+_33614214
|
0.541
|
|
ANXA2P2
|
annexin A2 pseudogene 2
|
|
chr7_-_104441285
|
0.541
|
|
LOC100216545
|
hypothetical LOC100216545
|
|
chr1_+_213807321
|
0.536
|
NM_016121
|
KCTD3
|
potassium channel tetramerisation domain containing 3
|
|
chr4_-_457839
|
0.535
|
|
ABCA11P
|
ATP-binding cassette, sub-family A (ABC1), member 11 (pseudogene)
|
|
chr7_-_112366962
|
0.533
|
NM_152556
|
C7orf60
|
chromosome 7 open reading frame 60
|
|
chr1_+_13948518
|
0.533
|
|
PRDM2
|
PR domain containing 2, with ZNF domain
|
|
chr15_+_75010997
|
0.532
|
NM_002902
|
RCN2
|
reticulocalbin 2, EF-hand calcium binding domain
|
|
chr17_-_45196516
|
0.532
|
NM_030802
|
FAM117A
|
family with sequence similarity 117, member A
|
|
chr5_+_133735004
|
0.530
|
|
UBE2B
|
ubiquitin-conjugating enzyme E2B (RAD6 homolog)
|
|
chr1_+_13948448
|
0.530
|
NM_001007257
|
PRDM2
|
PR domain containing 2, with ZNF domain
|
|
chr1_-_11245181
|
0.528
|
|
MTOR
|
mechanistic target of rapamycin (serine/threonine kinase)
|
|
chr20_+_33823318
|
0.528
|
NM_016436
|
PHF20
|
PHD finger protein 20
|
|
chr1_+_42619014
|
0.527
|
NM_173642
|
RIMKLA
|
ribosomal modification protein rimK-like family member A
|
|
chr15_-_98959898
|
0.524
|
NM_001040616
|
LINS
|
lines homolog (Drosophila)
|
|
chr8_-_103737058
|
0.523
|
NM_005655
|
KLF10
|
Kruppel-like factor 10
|
|
chr2_-_179380301
|
0.523
|
NM_003319
NM_133378
NM_133379
NM_133432
NM_133437
|
TTN
|
titin
|
|
chr1_-_7998265
|
0.520
|
|
ERRFI1
|
ERBB receptor feedback inhibitor 1
|
|
chr1_+_170768635
|
0.520
|
NM_014283
|
C1orf9
|
chromosome 1 open reading frame 9
|
|
chr4_+_160408447
|
0.519
|
NM_014247
|
RAPGEF2
|
Rap guanine nucleotide exchange factor (GEF) 2
|
|
chr10_-_112668679
|
0.519
|
NM_001195306
|
BBIP1
|
BBSome interacting protein 1
|
|
chr11_-_31487685
|
0.517
|
NM_144981
|
IMMP1L
|
IMP1 inner mitochondrial membrane peptidase-like (S. cerevisiae)
|
|
chr17_+_43539909
|
0.517
|
NM_152244
NM_013323
|
SNX11
|
sorting nexin 11
|
|
chr1_-_11245193
|
0.517
|
NM_004958
|
MTOR
|
mechanistic target of rapamycin (serine/threonine kinase)
|
|
chr1_-_23543330
|
0.517
|
NM_001102397
NM_001102398
NM_001102399
NM_005826
|
HNRNPR
|
heterogeneous nuclear ribonucleoprotein R
|
|
chr5_-_92982927
|
0.514
|
|
|
|
|
chr10_+_112669353
|
0.513
|
|
SHOC2
|
soc-2 suppressor of clear homolog (C. elegans)
|
|
chr5_-_58331515
|
0.513
|
NM_001197223
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr3_+_143078202
|
0.512
|
|
ATP1B3
|
ATPase, Na+/K+ transporting, beta 3 polypeptide
|
|
chr19_+_2818332
|
0.511
|
NM_024967
|
ZNF556
|
zinc finger protein 556
|
|
chr11_+_107497462
|
0.510
|
NM_000019
|
ACAT1
|
acetyl-CoA acetyltransferase 1
|
|
chr15_-_54544531
|
0.509
|
NM_018365
|
MNS1
|
meiosis-specific nuclear structural 1
|
|
chr11_-_129377929
|
0.509
|
NM_020228
NM_199437
|
PRDM10
|
PR domain containing 10
|
|
chr12_-_11136178
|
0.509
|
NM_176884
|
TAS2R43
|
taste receptor, type 2, member 43
|
|
chr12_+_27066749
|
0.508
|
NM_004264
|
MED21
|
mediator complex subunit 21
|
|
chr1_-_159257486
|
0.504
|
|
F11R
|
F11 receptor
|
|
chr17_-_45196465
|
0.502
|
|
FAM117A
|
family with sequence similarity 117, member A
|
|
chr2_+_61146866
|
0.502
|
NM_032506
|
KIAA1841
|
KIAA1841
|
|
chr5_-_27074407
|
0.501
|
NM_016279
|
CDH9
|
cadherin 9, type 2 (T1-cadherin)
|
|
chr5_-_150118274
|
0.501
|
NM_001135644
|
DCTN4
|
dynactin 4 (p62)
|
|
chr17_-_36181927
|
0.500
|
NM_181539
|
KRT26
|
keratin 26
|
|
chr5_+_179065282
|
0.499
|
|
CANX
|
calnexin
|