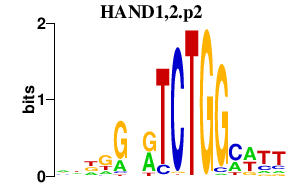
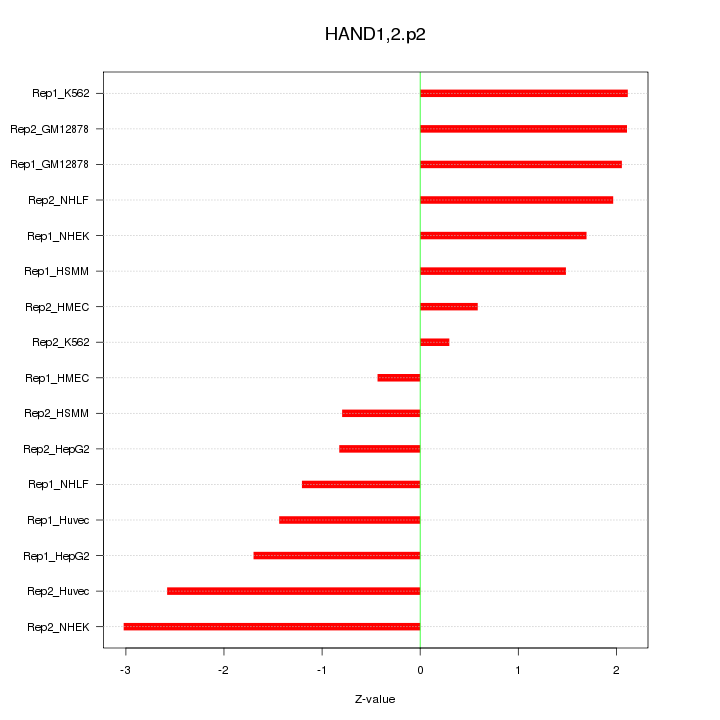
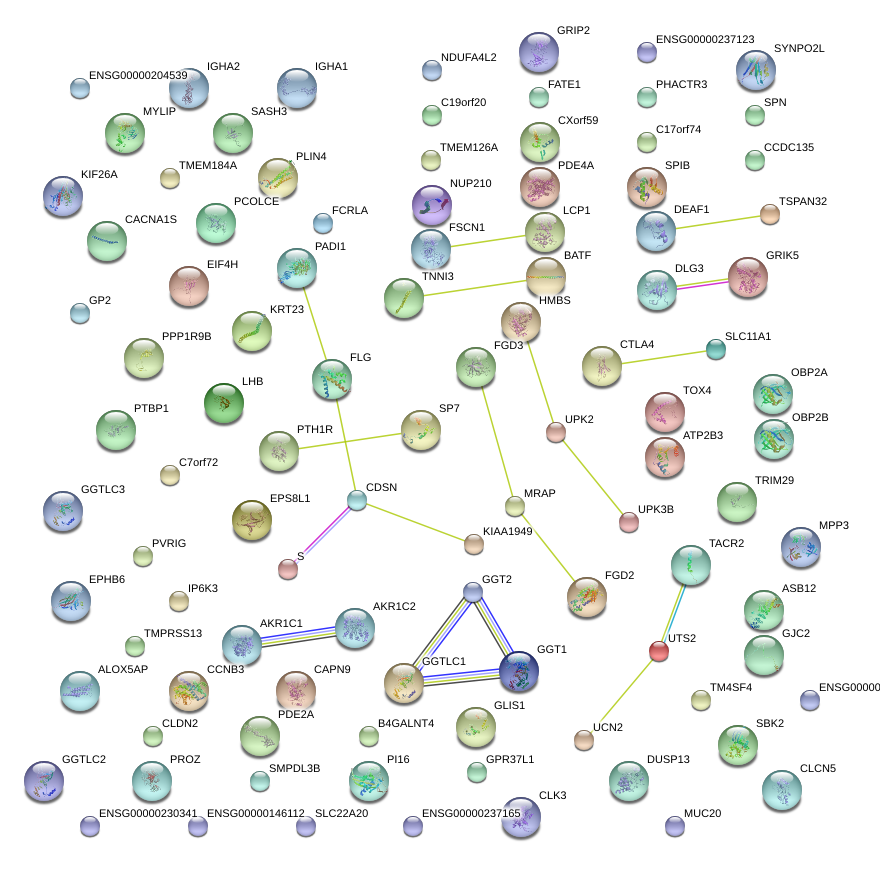
|
chr7_+_142270634
|
2.431
|
|
EPHB6
|
EPH receptor B6
|
|
chr19_+_55614006
|
2.426
|
NM_003121
|
SPIB
|
Spi-B transcription factor (Spi-1/PU.1 related)
|
|
chr1_+_557269
|
1.933
|
|
|
|
|
chr1_+_556968
|
1.925
|
|
|
|
|
chr12_+_119617060
|
1.918
|
|
|
|
|
chr2_+_204440753
|
1.697
|
NM_001037631
NM_005214
|
CTLA4
|
cytotoxic T-lymphocyte-associated protein 4
|
|
chr12_-_55917134
|
1.619
|
|
NDUFA4L2
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4-like 2
|
|
chr19_+_458475
|
1.505
|
NM_033513
|
C19orf20
|
chromosome 19 open reading frame 20
|
|
chr11_-_72063059
|
1.437
|
NM_002599
|
PDE2A
|
phosphodiesterase 2A, cGMP-stimulated
|
|
chr22_+_23333610
|
1.421
|
NM_001032365
|
GGT1
|
gamma-glutamyltransferase 1
|
|
chr7_+_99654806
|
1.411
|
NM_024070
|
PVRIG
|
poliovirus receptor related immunoglobulin domain containing
|
|
chr17_+_45617676
|
1.394
|
|
|
|
|
chr19_+_60279032
|
1.393
|
NM_133180
|
EPS8L1
|
EPS8-like 1
|
|
chr1_+_200358594
|
1.327
|
NM_004767
|
GPR37L1
|
G protein-coupled receptor 37 like 1
|
|
chrX_+_49718954
|
1.293
|
NM_000084
|
CLCN5
|
chloride channel 5
|
|
chr7_+_5609416
|
1.271
|
|
FSCN1
|
fascin homolog 1, actin-bundling protein (Strongylocentrotus purpuratus)
|
|
chr3_+_46894239
|
1.215
|
NM_000316
|
PTH1R
|
parathyroid hormone 1 receptor
|
|
chr11_-_117305277
|
1.211
|
NM_001077263
|
TMPRSS13
|
transmembrane protease, serine 13
|
|
chr13_-_45640679
|
1.173
|
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin)
|
|
chr11_+_359723
|
1.164
|
NM_178537
|
B4GALNT4
|
beta-1,4-N-acetyl-galactosaminyl transferase 4
|
|
chr6_+_37081387
|
1.154
|
NM_173558
|
FGD2
|
FYVE, RhoGEF and PH domain containing 2
|
|
chrX_+_50044279
|
1.150
|
NM_033031
NM_033670
|
CCNB3
|
cyclin B3
|
|
chr1_-_199348316
|
1.144
|
NM_000069
|
CACNA1S
|
calcium channel, voltage-dependent, L type, alpha 1S subunit
|
|
chr19_-_4468715
|
1.142
|
NM_001080400
|
PLIN4
|
perilipin 4
|
|
chr1_+_159943385
|
1.128
|
NM_001184866
NM_001184867
NM_001184870
NM_001184871
NM_001184872
NM_001184873
NM_032738
|
FCRLA
|
Fc receptor-like A
|
|
chr14_-_105392671
|
1.117
|
|
|
|
|
chr10_-_76529253
|
1.110
|
NM_016364
|
DUSP13
|
dual specificity phosphatase 13
|
|
chr7_-_1562272
|
1.078
|
NM_001097620
|
TMEM184A
|
transmembrane protein 184A
|
|
chr3_+_150675117
|
1.066
|
NM_004617
|
TM4SF4
|
transmembrane 4 L six family member 4
|
|
chr19_-_60360768
|
1.058
|
NM_000363
|
TNNI3
|
troponin I type 3 (cardiac)
|
|
chr19_+_750281
|
1.052
|
|
PTBP1
|
polypyrimidine tract binding protein 1
|
|
chr10_-_75080784
|
1.023
|
NM_024875
|
SYNPO2L
|
synaptopodin 2-like
|
|
chr1_-_53972464
|
1.019
|
NM_147193
|
GLIS1
|
GLIS family zinc finger 1
|
|
chrX_+_128741562
|
1.012
|
NM_018990
|
SASH3
|
SAM and SH3 domain containing 3
|
|
chr14_+_103675194
|
0.998
|
|
KIF26A
|
kinesin family member 26A
|
|
chr9_+_137577805
|
0.996
|
NM_014582
|
OBP2A
|
odorant binding protein 2A
|
|
chr19_-_47261761
|
0.993
|
NM_002088
|
GRIK5
|
glutamate receptor, ionotropic, kainate 5
|
|
chr2_+_218954995
|
0.971
|
NM_000578
|
SLC11A1
|
solute carrier family 11 (proton-coupled divalent metal ion transporters), member 1
|
|
chr12_-_52016270
|
0.967
|
NM_001173467
|
SP7
|
Sp7 transcription factor
|
|
chr20_+_57729659
|
0.962
|
NM_183244
NM_183246
|
PHACTR3
|
phosphatase and actin regulator 3
|
|
chrX_+_128741578
|
0.960
|
|
SASH3
|
SAM and SH3 domain containing 3
|
|
chrX_+_106030049
|
0.959
|
NM_001171092
|
CLDN2
|
claudin 2
|
|
chr19_-_60740246
|
0.956
|
NM_001101401
|
SBK2
|
SH3-binding domain kinase family, member 2
|
|
chrX_+_35974973
|
0.955
|
NM_173695
|
CXorf59
|
chromosome X open reading frame 59
|
|
chr5_+_148766632
|
0.941
|
|
LOC728264
|
hypothetical LOC728264
|
|
chr14_+_75058591
|
0.923
|
|
BATF
|
basic leucine zipper transcription factor, ATF-like
|
|
chr7_+_100037817
|
0.921
|
NM_002593
|
PCOLCE
|
procollagen C-endopeptidase enhancer
|
|
chr1_+_226404037
|
0.913
|
NM_020435
|
GJC2
|
gap junction protein, gamma 2, 47kDa
|
|
chr11_+_64737886
|
0.905
|
NM_001004326
|
SLC22A20
|
solute carrier family 22, member 20
|
|
chr3_-_14558591
|
0.895
|
NM_001080423
|
GRIP2
|
glutamate receptor interacting protein 2
|
|
chr10_-_5035967
|
0.883
|
NM_001135241
|
AKR1C1
AKR1C2
|
aldo-keto reductase family 1, member C1 (dihydrodiol dehydrogenase 1; 20-alpha (3-alpha)-hydroxysteroid dehydrogenase)
aldo-keto reductase family 1, member C2 (dihydrodiol dehydrogenase 2; bile acid binding protein; 3-alpha hydroxysteroid dehydrogenase, type III)
|
|
chr17_+_7269658
|
0.881
|
NM_175734
|
C17orf74
|
chromosome 17 open reading frame 74
|
|
chr7_+_75977680
|
0.874
|
NM_030570
NM_182684
|
UPK3B
|
uroplakin 3B
|
|
chr13_+_112860968
|
0.869
|
NM_003891
|
PROZ
|
protein Z, vitamin K-dependent plasma glycoprotein
|
|
chr16_+_56286212
|
0.868
|
NM_032269
|
CCDC135
|
coiled-coil domain containing 135
|
|
chr11_+_2279932
|
0.865
|
|
TSPAN32
|
tetraspanin 32
|
|
chr7_+_73248308
|
0.864
|
|
EIF4H
|
eukaryotic translation initiation factor 4H
|
|
chr16_-_20246309
|
0.861
|
NM_001007240
NM_001007241
NM_001007242
NM_001502
|
GP2
|
glycoprotein 2 (zymogen granule membrane)
|
|
chr14_+_75058518
|
0.858
|
NM_006399
|
BATF
|
basic leucine zipper transcription factor, ATF-like
|
|
chr3_+_196933422
|
0.857
|
NM_152673
|
MUC20
|
mucin 20, cell surface associated
|
|
chr6_-_33822659
|
0.854
|
NM_001142883
NM_054111
|
IP6K3
|
inositol hexakisphosphate kinase 3
|
|
chrX_-_63362227
|
0.850
|
NM_130388
|
ASB12
|
ankyrin repeat and SOCS box containing 12
|
|
chr6_-_30760413
|
0.849
|
|
KIAA1949
|
KIAA1949
|
|
chrX_+_69589081
|
0.847
|
|
DLG3
|
discs, large homolog 3 (Drosophila)
|
|
chr16_+_29582065
|
0.838
|
|
SPN
|
sialophorin
|
|
chr1_+_228949752
|
0.833
|
NM_006615
NM_016452
|
CAPN9
|
calpain 9
|
|
chr15_+_72687765
|
0.831
|
NM_003992
|
CLK3
|
CDC-like kinase 3
|
|
chr3_-_48576204
|
0.815
|
NM_033199
|
UCN2
|
urocortin 2
|
|
chr13_+_30207690
|
0.805
|
|
ALOX5AP
|
arachidonate 5-lipoxygenase-activating protein
|
|
chr11_-_119504726
|
0.804
|
|
TRIM29
|
tripartite motif containing 29
|
|
chr11_+_2279812
|
0.797
|
NM_139022
|
TSPAN32
|
tetraspanin 32
|
|
chr3_-_13359746
|
0.795
|
|
NUP210
|
nucleoporin 210kDa
|
|
chr7_+_50106227
|
0.789
|
NM_001161834
|
C7orf72
|
chromosome 7 open reading frame 72
|
|
chr11_+_118332217
|
0.783
|
NM_006760
|
UPK2
|
uroplakin 2
|
|
chr14_-_105845570
|
0.782
|
|
IGHA1
|
immunoglobulin heavy constant alpha 1
|
|
chr1_+_17404206
|
0.776
|
NM_013358
|
PADI1
|
peptidyl arginine deiminase, type I
|
|
chr1_+_16131633
|
0.766
|
|
|
|
|
chrX_+_152454773
|
0.746
|
NM_001001344
NM_021949
|
ATP2B3
|
ATPase, Ca++ transporting, plasma membrane 3
|
|
chr17_-_36346240
|
0.746
|
|
KRT23
|
keratin 23 (histone deacetylase inducible)
|
|
chr10_-_70846573
|
0.733
|
NM_001057
|
TACR2
|
tachykinin receptor 2
|
|
chr22_+_21053972
|
0.731
|
|
|
|
|
chr9_+_94776538
|
0.727
|
NM_033086
|
FGD3
|
FYVE, RhoGEF and PH domain containing 3
|
|
chr6_+_37030186
|
0.724
|
NM_153370
|
PI16
|
peptidase inhibitor 16
|
|
chr11_+_118463906
|
0.724
|
NM_001024382
|
HMBS
|
hydroxymethylbilane synthase
|
|
chrX_+_150635162
|
0.721
|
NM_033085
|
FATE1
|
fetal and adult testis expressed 1
|
|
chr6_-_31196174
|
0.718
|
NM_001264
|
CDSN
|
corneodesmosin
|
|
chr22_-_16127520
|
0.717
|
|
CECR3
|
cat eye syndrome chromosome region, candidate 3
|
|
chr1_+_28134090
|
0.712
|
NM_001009568
NM_014474
|
SMPDL3B
|
sphingomyelin phosphodiesterase, acid-like 3B
|
|
chr6_+_16251260
|
0.711
|
|
MYLIP
|
myosin regulatory light chain interacting protein
|
|
chr19_+_10404110
|
0.711
|
NM_001111309
|
PDE4A
|
phosphodiesterase 4A, cAMP-specific
|
|
chr19_-_54212158
|
0.710
|
NM_000894
|
LHB
|
luteinizing hormone beta polypeptide
|
|
chr12_-_120725210
|
0.709
|
|
LOC338799
|
hypothetical LOC338799
|
|
chr10_-_121286034
|
0.705
|
NM_002925
|
RGS10
|
regulator of G-protein signaling 10
|
|
chr11_-_2279642
|
0.704
|
NM_001142946
|
C11orf21
|
chromosome 11 open reading frame 21
|
|
chr8_-_19584373
|
0.703
|
NM_018371
|
CSGALNACT1
|
chondroitin sulfate N-acetylgalactosaminyltransferase 1
|
|
chr11_-_33139449
|
0.697
|
NM_001033505
NM_001033506
NM_001326
|
CSTF3
|
cleavage stimulation factor, 3' pre-RNA, subunit 3, 77kDa
|
|
chr6_+_167445284
|
0.692
|
NM_004367
|
CCR6
|
chemokine (C-C motif) receptor 6
|
|
chr10_+_82287637
|
0.689
|
NM_207372
|
SH2D4B
|
SH2 domain containing 4B
|
|
chr16_+_82885821
|
0.688
|
NM_021197
|
WFDC1
|
WAP four-disulfide core domain 1
|
|
chrX_-_69396241
|
0.681
|
NM_002565
|
P2RY4
|
pyrimidinergic receptor P2Y, G-protein coupled, 4
|
|
chr19_+_17047569
|
0.678
|
NM_001130065
NM_004145
|
MYO9B
|
myosin IXB
|
|
chr7_+_101978777
|
0.675
|
NM_001166339
NM_001031618
|
SPDYE2L
SPDYE2
|
WBSCR19-like protein 3
speedy homolog E2 (Xenopus laevis)
|
|
chr17_+_71235324
|
0.674
|
|
ITGB4
|
integrin, beta 4
|
|
chr12_-_52015696
|
0.671
|
NM_152860
|
SP7
|
Sp7 transcription factor
|
|
chr11_+_2439241
|
0.662
|
NM_181798
|
KCNQ1
|
potassium voltage-gated channel, KQT-like subfamily, member 1
|
|
chr1_-_29322993
|
0.660
|
NM_001003682
|
TMEM200B
|
transmembrane protein 200B
|
|
chrX_-_106033202
|
0.655
|
NM_001171706
NM_138382
|
RIPPLY1
|
ripply1 homolog (zebrafish)
|
|
chr6_-_133161318
|
0.646
|
NM_052831
|
C6orf192
|
chromosome 6 open reading frame 192
|
|
chr14_-_64508547
|
0.636
|
NM_198686
|
RAB15
|
RAB15, member RAS onocogene family
|
|
chr11_+_66927976
|
0.634
|
NM_198517
|
TBC1D10C
|
TBC1 domain family, member 10C
|
|
chr17_-_76897590
|
0.633
|
NM_178519
|
C17orf55
|
chromosome 17 open reading frame 55
|
|
chr16_+_12053555
|
0.630
|
NM_001080530
|
SNX29
|
sorting nexin 29
|
|
chr17_+_35038276
|
0.622
|
NM_181505
|
PPP1R1B
|
protein phosphatase 1, regulatory (inhibitor) subunit 1B
|
|
chr1_+_32433605
|
0.612
|
|
TXLNA
|
taxilin alpha
|
|
chr1_+_199519202
|
0.612
|
NM_000299
NM_001005337
|
PKP1
|
plakophilin 1 (ectodermal dysplasia/skin fragility syndrome)
|
|
chr16_+_28413421
|
0.611
|
NM_018690
|
APOBR
|
apolipoprotein B receptor
|
|
chr3_+_150675169
|
0.611
|
|
TM4SF4
|
transmembrane 4 L six family member 4
|
|
chrX_-_100759562
|
0.608
|
NM_001009584
NM_001184768
NM_019007
|
ARMCX6
|
armadillo repeat containing, X-linked 6
|
|
chr13_+_30207668
|
0.606
|
NM_001629
|
ALOX5AP
|
arachidonate 5-lipoxygenase-activating protein
|
|
chr7_-_100630912
|
0.602
|
NM_178176
|
MOGAT3
|
monoacylglycerol O-acyltransferase 3
|
|
chr6_-_39390073
|
0.601
|
NM_031460
NM_001135111
|
KCNK17
|
potassium channel, subfamily K, member 17
|
|
chr21_+_41616590
|
0.600
|
|
FAM3B
|
family with sequence similarity 3, member B
|
|
chr21_-_42644138
|
0.600
|
NM_005423
|
TFF2
|
trefoil factor 2
|
|
chr19_+_5865192
|
0.600
|
NM_004058
NM_080590
|
CAPS
|
calcyphosine
|
|
chr9_-_35744271
|
0.599
|
NM_001044264
|
MSMP
|
microseminoprotein, prostate associated
|
|
chrX_+_130505630
|
0.595
|
NM_001004486
|
OR13H1
|
olfactory receptor, family 13, subfamily H, member 1
|
|
chr10_+_118946989
|
0.591
|
NM_181840
|
KCNK18
|
potassium channel, subfamily K, member 18
|
|
chr15_+_67393795
|
0.590
|
NM_001104554
|
PAQR5
|
progestin and adipoQ receptor family member V
|
|
chr1_+_46418786
|
0.588
|
|
TSPAN1
|
tetraspanin 1
|
|
chr12_-_108755588
|
0.585
|
NM_021625
|
TRPV4
|
transient receptor potential cation channel, subfamily V, member 4
|
|
chr1_-_203868622
|
0.570
|
NM_001973
NM_021795
|
ELK4
|
ELK4, ETS-domain protein (SRF accessory protein 1)
|
|
chr17_+_14145051
|
0.570
|
NM_006041
|
HS3ST3B1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 3B1
|
|
chr5_+_36644187
|
0.566
|
NM_001166695
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3
|
|
chr1_+_152560179
|
0.557
|
NM_080429
|
AQP10
|
aquaporin 10
|
|
chr2_+_128119908
|
0.555
|
NM_001161415
NM_001161416
NM_001161417
NM_005291
|
GPR17
|
G protein-coupled receptor 17
|
|
chr21_+_43024936
|
0.550
|
|
PDE9A
|
phosphodiesterase 9A
|
|
chr3_+_196933481
|
0.548
|
|
MUC20
|
mucin 20, cell surface associated
|
|
chr9_-_131555111
|
0.548
|
NM_004878
|
PTGES
|
prostaglandin E synthase
|
|
chr5_+_38439399
|
0.546
|
NM_182798
NM_182799
|
EGFLAM
|
EGF-like, fibronectin type III and laminin G domains
|
|
chr19_+_11510578
|
0.543
|
NM_001299
|
CNN1
|
calponin 1, basic, smooth muscle
|
|
chrX_+_150653873
|
0.542
|
NM_005140
|
CNGA2
|
cyclic nucleotide gated channel alpha 2
|
|
chr7_-_142293382
|
0.535
|
NM_018646
|
TRPV6
|
transient receptor potential cation channel, subfamily V, member 6
|
|
chrX_-_10605726
|
0.535
|
NM_001098624
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chr11_+_1420250
|
0.535
|
|
BRSK2
|
BR serine/threonine kinase 2
|
|
chrX_+_129363575
|
0.534
|
NM_016024
|
RBMX2
|
RNA binding motif protein, X-linked 2
|
|
chr7_-_150385912
|
0.529
|
NM_001164410
NM_004935
|
CDK5
|
cyclin-dependent kinase 5
|
|
chr16_+_639278
|
0.528
|
NM_145294
|
WDR90
|
WD repeat domain 90
|
|
chr19_+_60879802
|
0.527
|
NM_001130071
|
EPN1
|
epsin 1
|
|
chr6_-_86407875
|
0.525
|
NM_001159675
NM_001159676
|
SYNCRIP
|
synaptotagmin binding, cytoplasmic RNA interacting protein
|
|
chr1_-_110949867
|
0.524
|
NM_004974
|
KCNA2
|
potassium voltage-gated channel, shaker-related subfamily, member 2
|
|
chr16_+_55612125
|
0.523
|
|
NLRC5
|
NLR family, CARD domain containing 5
|
|
chr19_+_10424581
|
0.521
|
NM_006202
|
PDE4A
|
phosphodiesterase 4A, cAMP-specific
|
|
chr12_+_1799693
|
0.520
|
NM_001039029
NM_001163925
NM_001163926
|
LRTM2
|
leucine-rich repeats and transmembrane domains 2
|
|
chr20_-_48686832
|
0.520
|
NM_080829
|
FAM65C
|
family with sequence similarity 65, member C
|
|
chr5_+_150008030
|
0.515
|
|
SYNPO
|
synaptopodin
|
|
chr17_-_9864048
|
0.515
|
|
GAS7
|
growth arrest-specific 7
|
|
chr17_-_37590910
|
0.515
|
NM_001524
|
HCRT
|
hypocretin (orexin) neuropeptide precursor
|
|
chr1_+_148511806
|
0.513
|
NM_024579
|
C1orf54
|
chromosome 1 open reading frame 54
|
|
chr9_+_135389698
|
0.511
|
NM_014694
|
ADAMTSL2
|
ADAMTS-like 2
|
|
chr1_+_26358097
|
0.511
|
NM_024869
|
GRRP1
|
glycine/arginine rich protein 1
|
|
chr11_-_64283945
|
0.504
|
|
PYGM
|
phosphorylase, glycogen, muscle
|
|
chr14_-_105163301
|
0.502
|
|
IGHG4
|
immunoglobulin heavy constant gamma 4 (G4m marker)
|
|
chr17_+_71232370
|
0.501
|
NM_001005619
|
ITGB4
|
integrin, beta 4
|
|
chr19_+_40511909
|
0.498
|
NM_001185099
NM_001185100
NM_001185101
NM_001771
|
CD22
|
CD22 molecule
|
|
chr9_-_33455191
|
0.497
|
|
NOL6
|
nucleolar protein family 6 (RNA-associated)
|
|
chr22_+_38296703
|
0.487
|
NM_001003406
NM_021096
|
CACNA1I
|
calcium channel, voltage-dependent, T type, alpha 1I subunit
|
|
chr11_+_123315459
|
0.487
|
NM_001001965
|
OR4D5
|
olfactory receptor, family 4, subfamily D, member 5
|
|
chr1_-_154041393
|
0.484
|
|
GON4L
|
gon-4-like (C. elegans)
|
|
chr19_+_44838373
|
0.479
|
NM_001190441
|
LGALS16
|
beta-galactoside-binding lectin
|
|
chrX_+_46935035
|
0.478
|
NM_153280
|
UBA1
|
ubiquitin-like modifier activating enzyme 1
|
|
chr20_-_62052922
|
0.477
|
NM_001193379
|
UCKL1
|
uridine-cytidine kinase 1-like 1
|
|
chr19_-_59542197
|
0.477
|
NM_012276
|
LILRA4
|
leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 4
|
|
chr12_-_108755565
|
0.476
|
|
TRPV4
|
transient receptor potential cation channel, subfamily V, member 4
|
|
chr3_+_42675841
|
0.474
|
|
ZBTB47
|
zinc finger and BTB domain containing 47
|
|
chr15_+_38430517
|
0.472
|
NM_001145643
|
PHGR1
|
proline/histidine/glycine-rich 1
|
|
chr8_-_27906287
|
0.471
|
NM_173833
|
SCARA5
|
scavenger receptor class A, member 5 (putative)
|
|
chr17_+_20423628
|
0.468
|
NM_001190790
|
CDRT15L2
|
CMT1A duplicated region transcript 15-like 2
|
|
chr18_-_58946858
|
0.466
|
|
BCL2
|
B-cell CLL/lymphoma 2
|
|
chr5_+_149867866
|
0.466
|
NM_001543
|
NDST1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1
|
|
chr14_-_87806913
|
0.463
|
NM_138318
|
KCNK10
|
potassium channel, subfamily K, member 10
|
|
chr11_-_70350292
|
0.462
|
|
SHANK2
|
SH3 and multiple ankyrin repeat domains 2
|
|
chr22_-_21814208
|
0.459
|
NM_014433
|
RTDR1
|
rhabdoid tumor deletion region gene 1
|
|
chr17_-_9863929
|
0.459
|
|
GAS7
|
growth arrest-specific 7
|
|
chr17_+_70984173
|
0.457
|
|
KIAA0195
|
KIAA0195
|
|
chr7_+_102078007
|
0.456
|
NM_001166339
NM_001031618
|
SPDYE2L
SPDYE2
|
WBSCR19-like protein 3
speedy homolog E2 (Xenopus laevis)
|
|
chr7_-_150385867
|
0.452
|
|
CDK5
|
cyclin-dependent kinase 5
|
|
chr11_+_22316242
|
0.452
|
NM_020346
|
SLC17A6
|
solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 6
|
|
chr17_+_6288457
|
0.451
|
NM_001195228
NM_019013
|
FAM64A
|
family with sequence similarity 64, member A
|
|
chr10_+_72642274
|
0.449
|
NM_170744
|
UNC5B
|
unc-5 homolog B (C. elegans)
|
|
chr6_+_31016755
|
0.445
|
NM_080870
|
DPCR1
|
diffuse panbronchiolitis critical region 1
|
|
chr17_+_43263991
|
0.441
|
NM_033413
|
LRRC46
|
leucine rich repeat containing 46
|
|
chr20_+_36643251
|
0.439
|
NM_001018082
|
ADIG
|
adipogenin
|
|
chrX_+_37735013
|
0.438
|
NM_012274
|
CXorf27
|
chromosome X open reading frame 27
|
|
chr3_+_38322430
|
0.437
|
NM_004803
|
SLC22A14
|
solute carrier family 22, member 14
|
|
chr22_-_27787793
|
0.435
|
NM_015370
|
C22orf31
|
chromosome 22 open reading frame 31
|
|
chr15_+_40484249
|
0.435
|
NM_173089
NM_173090
|
CAPN3
|
calpain 3, (p94)
|
|
chr15_+_52092392
|
0.429
|
NM_001080534
|
UNC13C
|
unc-13 homolog C (C. elegans)
|
|
chr1_-_30977931
|
0.426
|
|
LAPTM5
|
lysosomal protein transmembrane 5
|
|
chr6_-_46997579
|
0.426
|
NM_001098518
|
GPR116
|
G protein-coupled receptor 116
|
|
chr12_-_50502474
|
0.425
|
NM_001013690
|
FIGNL2
|
fidgetin-like 2
|