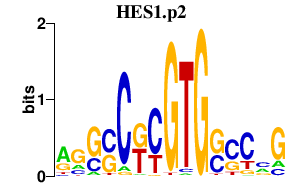
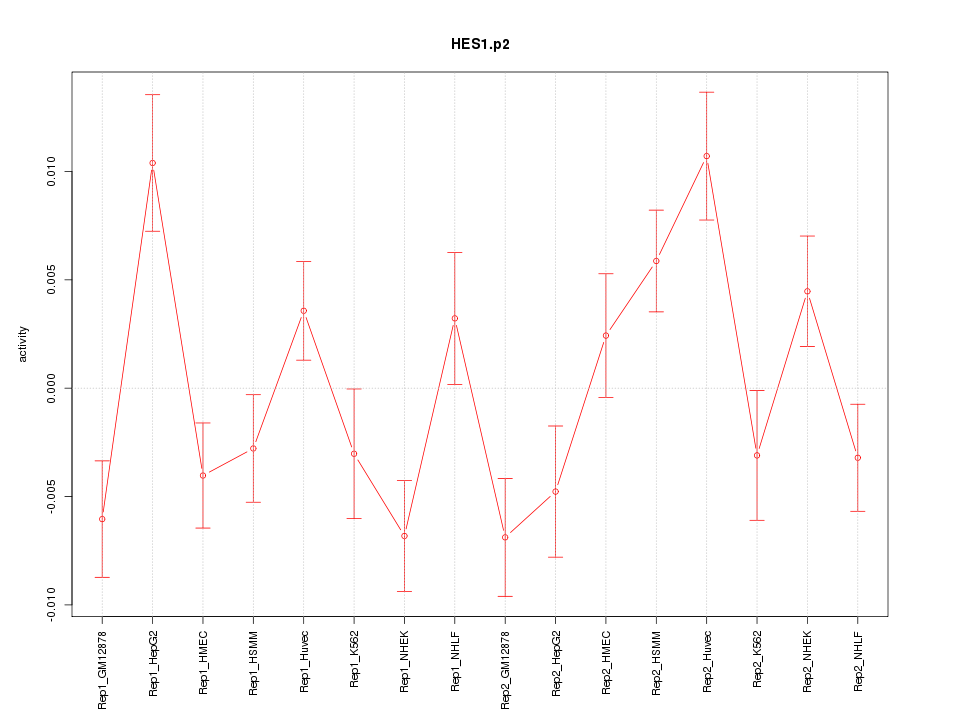
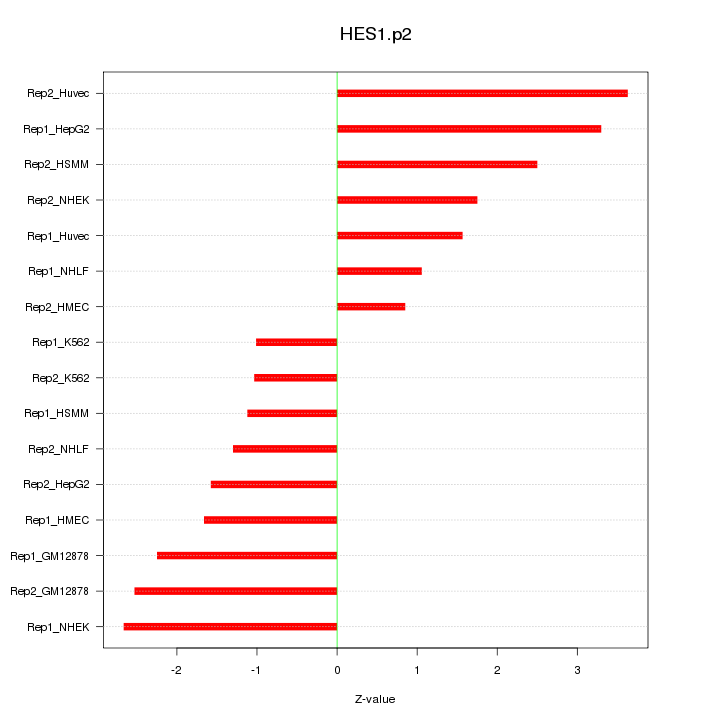
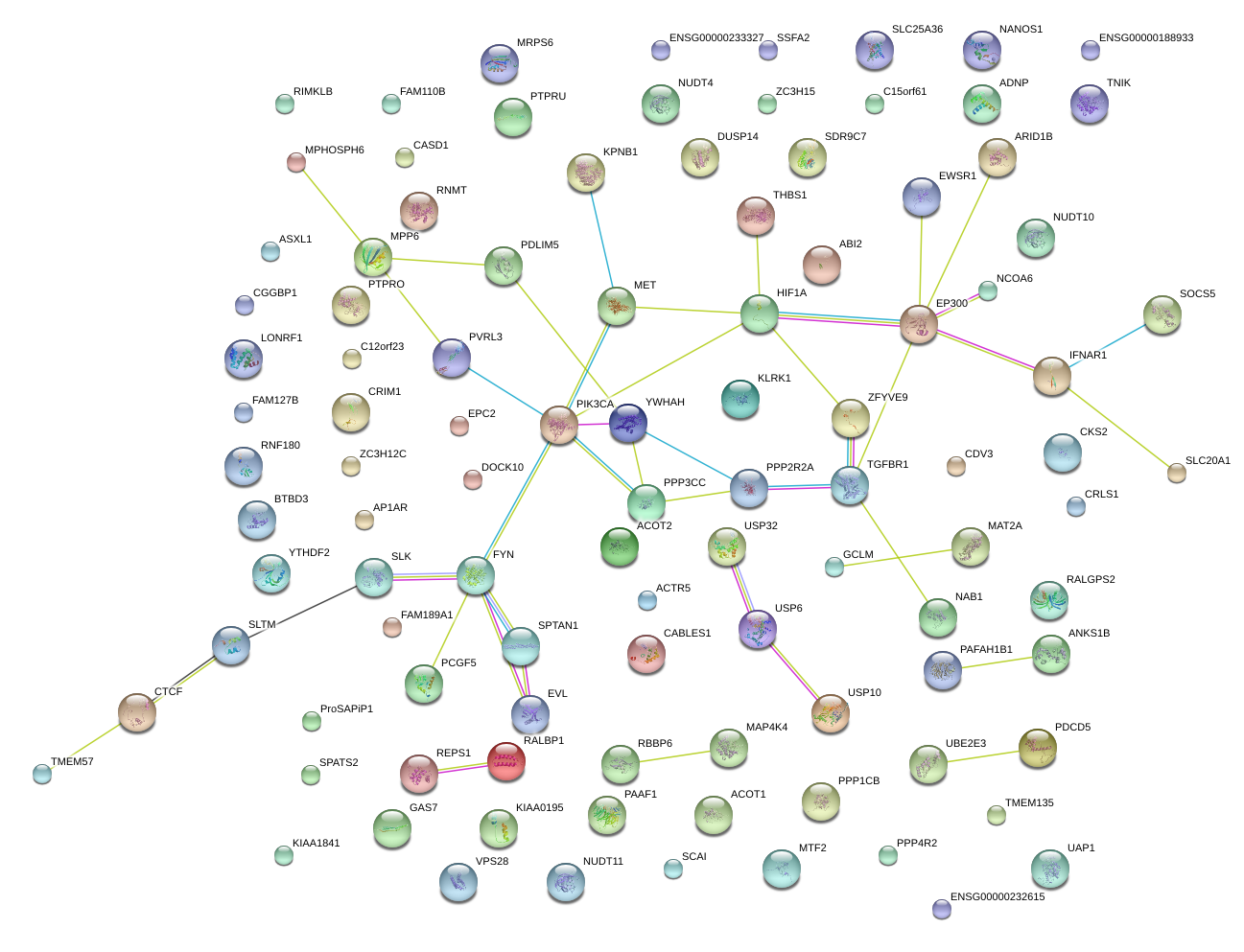
|
chr20_+_11819364
|
5.139
|
NM_181443
|
BTBD3
|
BTB (POZ) domain containing 3
|
|
chr2_+_113119985
|
2.354
|
NM_005415
|
SLC20A1
|
solute carrier family 20 (phosphate transporter), member 1
|
|
chr12_+_8741746
|
2.353
|
NM_020734
|
RIMKLB
|
ribosomal modification protein rimK-like family member B
|
|
chr2_+_46779827
|
2.302
|
NM_144949
|
SOCS5
|
suppressor of cytokine signaling 5
|
|
chr14_+_61231795
|
2.152
|
NM_001530
NM_181054
|
HIF1A
|
hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor)
|
|
chr2_+_182464910
|
2.089
|
|
SSFA2
|
sperm specific antigen 2
|
|
chr3_-_88190737
|
2.078
|
NM_001008390
NM_003663
|
CGGBP1
|
CGG triplet repeat binding protein 1
|
|
chr3_+_112273464
|
1.925
|
NM_015480
|
PVRL3
|
poliovirus receptor-related 3
|
|
chr7_+_93977267
|
1.897
|
|
CASD1
|
CAS1 domain containing 1
|
|
chr17_+_43082273
|
1.874
|
NM_002265
|
KPNB1
|
karyopherin (importin) beta 1
|
|
chr22_+_39817733
|
1.848
|
|
EP300
|
E1A binding protein p300
|
|
chr2_+_181553613
|
1.824
|
|
UBE2E3
|
ubiquitin-conjugating enzyme E2E 3 (UBC4/5 homolog, yeast)
|
|
chr2_-_225615556
|
1.761
|
NM_014689
|
DOCK10
|
dedicator of cytokinesis 10
|
|
chr2_+_85619802
|
1.754
|
|
MAT2A
|
methionine adenosyltransferase II, alpha
|
|
chr5_+_63497408
|
1.743
|
NM_001113561
NM_178532
|
RNF180
|
ring finger protein 180
|
|
chr2_+_182464713
|
1.729
|
NM_001130445
NM_006751
|
SSFA2
|
sperm specific antigen 2
|
|
chr8_-_12657339
|
1.709
|
NM_152271
|
LONRF1
|
LON peptidase N-terminal domain and ring finger 1
|
|
chr3_+_180348818
|
1.535
|
NM_006218
|
PIK3CA
|
phosphoinositide-3-kinase, catalytic, alpha polypeptide
|
|
chr8_+_59069522
|
1.532
|
NM_147189
|
FAM110B
|
family with sequence similarity 110, member B
|
|
chr7_+_24579239
|
1.453
|
NM_016447
|
MPP6
|
membrane protein, palmitoylated 6 (MAGUK p55 subfamily member 6)
|
|
chr11_+_86426518
|
1.448
|
NM_001168724
NM_022918
|
TMEM135
|
transmembrane protein 135
|
|
chr21_+_34367747
|
1.437
|
|
MRPS6
|
mitochondrial ribosomal protein S6
|
|
chr2_+_101680594
|
1.419
|
NM_004834
NM_145686
NM_145687
|
MAP4K4
|
mitogen-activated protein kinase kinase kinase kinase 4
|
|
chr17_-_10042579
|
1.373
|
NM_201433
|
GAS7
|
growth arrest-specific 7
|
|
chr9_+_100907189
|
1.366
|
|
TGFBR1
|
transforming growth factor, beta receptor 1
|
|
chr17_+_43081837
|
1.337
|
|
KPNB1
|
karyopherin (importin) beta 1
|
|
chr9_+_100907222
|
1.334
|
NM_001130916
NM_004612
|
TGFBR1
|
transforming growth factor, beta receptor 1
|
|
chr10_+_120779217
|
1.331
|
NM_199461
|
NANOS1
|
nanos homolog 1 (Drosophila)
|
|
chr9_+_100907213
|
1.316
|
|
TGFBR1
|
transforming growth factor, beta receptor 1
|
|
chr17_+_43082302
|
1.306
|
|
KPNB1
|
karyopherin (importin) beta 1
|
|
chr2_+_61146866
|
1.302
|
NM_032506
|
KIAA1841
|
KIAA1841
|
|
chr3_+_73128771
|
1.276
|
NM_174907
|
PPP4R2
|
protein phosphatase 4, regulatory subunit 2
|
|
chr2_+_203901160
|
1.272
|
NM_005759
|
ABI2
|
abl-interactor 2
|
|
chr7_+_93976927
|
1.272
|
NM_022900
|
CASD1
|
CAS1 domain containing 1
|
|
chr9_+_100907292
|
1.254
|
|
|
|
|
chr22_+_30670435
|
1.234
|
NM_003405
|
YWHAH
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta polypeptide
|
|
chr18_+_18969724
|
1.226
|
NM_001100619
|
CABLES1
|
Cdk5 and Abl enzyme substrate 1
|
|
chr3_+_134775404
|
1.226
|
|
CDV3
|
CDV3 homolog (mouse)
|
|
chr4_+_119419354
|
1.221
|
|
SNHG8
|
small nucleolar RNA host gene 8 (non-protein coding)
|
|
chr2_+_181554048
|
1.206
|
|
UBE2E3
|
ubiquitin-conjugating enzyme E2E 3 (UBC4/5 homolog, yeast)
|
|
chr14_+_99507848
|
1.197
|
|
EVL
|
Enah/Vasp-like
|
|
chr16_+_24458349
|
1.168
|
NM_006910
NM_018703
NM_032626
|
RBBP6
|
retinoblastoma binding protein 6
|
|
chr1_+_93317374
|
1.165
|
NM_001164391
NM_001164392
NM_001164393
NM_007358
|
MTF2
|
metal response element binding transcription factor 2
|
|
chr9_-_126945536
|
1.164
|
NM_001144877
NM_173690
|
SCAI
|
suppressor of cancer cell invasion
|
|
chr6_-_86445138
|
1.151
|
|
SNHG5
|
small nucleolar RNA host gene 5 (non-protein coding)
|
|
chr10_+_92912676
|
1.144
|
|
PCGF5
|
polycomb group ring finger 5
|
|
chr9_+_130354490
|
1.137
|
NM_001130438
NM_001195532
NM_003127
|
SPTAN1
|
spectrin, alpha, non-erythrocytic 1 (alpha-fodrin)
|
|
chr20_-_32877029
|
1.137
|
NM_014071
|
NCOA6
|
nuclear receptor coactivator 6
|
|
chr15_+_65600520
|
1.121
|
NM_001143936
|
C15orf61
|
chromosome 15 open reading frame 61
|
|
chr22_+_30670543
|
1.115
|
|
YWHAH
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta polypeptide
|
|
chr1_+_176960896
|
1.112
|
NM_152663
|
RALGPS2
|
Ral GEF with PH domain and SH3 binding motif 2
|
|
chr1_-_94147537
|
1.089
|
|
GCLM
|
glutamate-cysteine ligase, modifier subunit
|
|
chr3_+_112273352
|
1.073
|
|
PVRL3
|
poliovirus receptor-related 3
|
|
chr6_-_163754903
|
1.068
|
|
LOC100526820
|
hypothetical LOC100526820
|
|
chr2_+_191222077
|
1.066
|
NM_005966
|
NAB1
|
NGFI-A binding protein 1 (EGR1 binding protein 1)
|
|
chr7_+_116099648
|
1.061
|
NM_000245
NM_001127500
|
MET
|
met proto-oncogene (hepatocyte growth factor receptor)
|
|
chr20_+_36810510
|
1.059
|
NM_024855
|
ACTR5
|
ARP5 actin-related protein 5 homolog (yeast)
|
|
chr21_+_33619067
|
1.053
|
NM_000629
|
IFNAR1
|
interferon (alpha, beta and omega) receptor 1
|
|
chr10_+_105716836
|
1.046
|
|
SLK
|
STE20-like kinase
|
|
chr12_-_98902562
|
1.042
|
NM_152788
|
ANKS1B
|
ankyrin repeat and sterile alpha motif domain containing 1B
|
|
chr1_-_94147574
|
1.039
|
NM_002061
|
GCLM
|
glutamate-cysteine ligase, modifier subunit
|
|
chr1_+_25629968
|
1.035
|
NM_018202
|
TMEM57
|
transmembrane protein 57
|
|
chr2_+_36436873
|
1.035
|
NM_016441
|
CRIM1
|
cysteine rich transmembrane BMP regulator 1 (chordin-like)
|
|
chr14_+_73073570
|
1.029
|
NM_001037161
|
ACOT1
|
acyl-CoA thioesterase 1
|
|
chr2_+_149118791
|
1.028
|
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila)
|
|
chr20_-_3102169
|
1.027
|
|
ProSAPiP1
|
ProSAPiP1 protein
|
|
chr18_+_9465528
|
1.022
|
NM_006788
|
RALBP1
|
ralA binding protein 1
|
|
chr6_+_157140733
|
1.019
|
NM_017519
NM_020732
|
ARID1B
|
AT rich interactive domain 1B (SWI1-like)
|
|
chr21_+_33619147
|
1.018
|
|
IFNAR1
|
interferon (alpha, beta and omega) receptor 1
|
|
chr11_+_109469296
|
1.013
|
NM_033390
|
ZC3H12C
|
zinc finger CCCH-type containing 12C
|
|
chr16_+_66153810
|
1.012
|
NM_001191022
NM_006565
|
CTCF
|
CCCTC-binding factor (zinc finger protein)
|
|
chr15_+_37660568
|
1.007
|
NM_003246
|
THBS1
|
thrombospondin 1
|
|
chr2_+_181553333
|
1.006
|
NM_182678
NM_006357
|
UBE2E3
|
ubiquitin-conjugating enzyme E2E 3 (UBC4/5 homolog, yeast)
|
|
chr20_+_30410447
|
1.004
|
|
ASXL1
|
additional sex combs like 1 (Drosophila)
|
|
chr16_+_83291055
|
1.002
|
NM_005153
|
USP10
|
ubiquitin specific peptidase 10
|
|
chr9_+_91115924
|
1.000
|
NM_001827
|
CKS2
|
CDC28 protein kinase regulatory subunit 2
|
|
chr2_+_85619797
|
0.998
|
NM_005911
|
MAT2A
|
methionine adenosyltransferase II, alpha
|
|
chr2_+_187059277
|
0.994
|
|
ZC3H15
|
zinc finger CCCH-type containing 15
|
|
chr15_-_27650218
|
0.993
|
NM_015307
|
FAM189A1
|
family with sequence similarity 189, member A1
|
|
chr2_+_36436316
|
0.992
|
|
CRIM1
|
cysteine rich transmembrane BMP regulator 1 (chordin-like)
|
|
chr20_-_48980858
|
0.991
|
NM_015339
NM_181442
|
ADNP
|
activity-dependent neuroprotector homeobox
|
|
chr1_+_52380633
|
0.991
|
NM_004799
NM_007323
NM_007324
|
ZFYVE9
|
zinc finger, FYVE domain containing 9
|
|
chr8_+_22354712
|
0.988
|
|
PPP3CC
|
protein phosphatase 3, catalytic subunit, gamma isozyme
|
|
chr17_+_2443652
|
0.987
|
NM_000430
|
PAFAH1B1
|
platelet-activating factor acetylhydrolase 1b, regulatory subunit 1 (45kDa)
|
|
chr12_+_48046946
|
0.984
|
NM_023071
|
SPATS2
|
spermatogenesis associated, serine-rich 2
|
|
chr8_+_22354532
|
0.983
|
NM_005605
|
PPP3CC
|
protein phosphatase 3, catalytic subunit, gamma isozyme
|
|
chr22_+_27993997
|
0.983
|
NM_001163285
NM_001163286
NM_001163287
NM_005243
NM_013986
|
EWSR1
|
Ewing sarcoma breakpoint region 1
|
|
chr3_-_189354510
|
0.957
|
|
LOC339929
|
hypothetical LOC339929
|
|
chr10_+_105716925
|
0.952
|
|
SLK
|
STE20-like kinase
|
|
chr4_+_95592067
|
0.951
|
|
PDLIM5
|
PDZ and LIM domain 5
|
|
chr16_+_83291097
|
0.936
|
|
USP10
|
ubiquitin specific peptidase 10
|
|
chr12_+_105873669
|
0.928
|
NM_152261
|
C12orf23
|
chromosome 12 open reading frame 23
|
|
chr4_+_95592030
|
0.925
|
NM_001011513
NM_001011515
NM_001011516
NM_006457
|
PDLIM5
|
PDZ and LIM domain 5
|
|
chr20_+_5934761
|
0.921
|
|
CRLS1
|
cardiolipin synthase 1
|
|
chr8_+_26204960
|
0.920
|
|
PPP2R2A
|
protein phosphatase 2, regulatory subunit B, alpha
|
|
chr3_+_142143329
|
0.916
|
NM_001104647
NM_018155
|
SLC25A36
|
solute carrier family 25, member 36
|
|
chr4_+_113372287
|
0.912
|
NM_001128426
NM_018569
|
AP1AR
|
adaptor-related protein complex 1 associated regulatory protein
|
|
chr12_+_15366712
|
0.910
|
NM_002848
NM_030667
|
PTPRO
|
protein tyrosine phosphatase, receptor type, O
|
|
chr2_+_28828112
|
0.899
|
NM_206876
NM_002709
|
PPP1CB
|
protein phosphatase 1, catalytic subunit, beta isozyme
|
|
chr20_+_5934560
|
0.893
|
NM_019095
|
CRLS1
|
cardiolipin synthase 1
|
|
chr6_+_157140895
|
0.893
|
|
|
|
|
chr8_+_26204997
|
0.888
|
|
PPP2R2A
|
protein phosphatase 2, regulatory subunit B, alpha
|
|
chr17_+_70964258
|
0.888
|
NM_014738
|
KIAA0195
|
KIAA0195
|
|
chr19_+_37763927
|
0.887
|
NM_004708
|
PDCD5
|
programmed cell death 5
|
|
chr1_+_160797973
|
0.883
|
|
UAP1
|
UDP-N-acteylglucosamine pyrophosphorylase 1
|
|
chr1_+_28936030
|
0.878
|
NM_001172828
|
YTHDF2
|
YTH domain family, member 2
|
|
chr17_+_32924110
|
0.877
|
|
DUSP14
|
dual specificity phosphatase 14
|
|
chr12_+_92295738
|
0.874
|
NM_019094
NM_199040
|
NUDT4
NUDT4P1
|
nudix (nucleoside diphosphate linked moiety X)-type motif 4
nudix (nucleoside diphosphate linked moiety X)-type motif 4 pseudogene 1
|
|
chr13_+_34414390
|
0.867
|
NM_015678
|
NBEA
|
neurobeachin
|
|
chr20_+_56899904
|
0.867
|
|
GNAS
|
GNAS complex locus
|
|
chr2_-_203444468
|
0.863
|
NM_138468
|
ICA1L
|
islet cell autoantigen 1,69kDa-like
|
|
chr2_+_200484251
|
0.863
|
|
C2orf69
|
chromosome 2 open reading frame 69
|
|
chr1_-_92537087
|
0.855
|
NM_053274
|
GLMN
|
glomulin, FKBP associated protein
|
|
chr13_-_109236897
|
0.850
|
NM_003749
|
IRS2
|
insulin receptor substrate 2
|
|
chr4_+_57469122
|
0.837
|
|
REST
|
RE1-silencing transcription factor
|
|
chr21_+_34367678
|
0.837
|
NM_032476
NM_006933
|
MRPS6
SLC5A3
|
mitochondrial ribosomal protein S6
solute carrier family 5 (sodium/myo-inositol cotransporter), member 3
|
|
chr2_+_54536780
|
0.833
|
NM_003128
|
SPTBN1
|
spectrin, beta, non-erythrocytic 1
|
|
chr1_+_92537109
|
0.833
|
NM_024813
|
RPAP2
|
RNA polymerase II associated protein 2
|
|
chr4_+_57468847
|
0.833
|
|
REST
|
RE1-silencing transcription factor
|
|
chr2_+_200484270
|
0.829
|
|
C2orf69
|
chromosome 2 open reading frame 69
|
|
chr5_+_14196287
|
0.828
|
NM_007118
|
TRIO
|
triple functional domain (PTPRF interacting)
|
|
chr15_+_83092854
|
0.819
|
|
ZNF592
|
zinc finger protein 592
|
|
chr8_+_87424074
|
0.812
|
NM_007013
|
WWP1
|
WW domain containing E3 ubiquitin protein ligase 1
|
|
chr3_+_189354167
|
0.809
|
NM_001167672
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr18_-_72336133
|
0.807
|
NM_014643
|
ZNF516
|
zinc finger protein 516
|
|
chr13_-_105985313
|
0.806
|
NM_004093
|
EFNB2
|
ephrin-B2
|
|
chr10_+_92970302
|
0.805
|
NM_032373
|
PCGF5
|
polycomb group ring finger 5
|
|
chr2_-_200424140
|
0.801
|
|
LOC348751
|
hypothetical LOC348751
|
|
chr13_+_51056648
|
0.800
|
|
WDFY2
|
WD repeat and FYVE domain containing 2
|
|
chr16_+_83291112
|
0.799
|
|
USP10
|
ubiquitin specific peptidase 10
|
|
chr15_+_38013559
|
0.795
|
NM_001013703
|
EIF2AK4
|
eukaryotic translation initiation factor 2 alpha kinase 4
|
|
chr3_+_184898234
|
0.794
|
NM_018023
|
YEATS2
|
YEATS domain containing 2
|
|
chr2_+_170298700
|
0.793
|
|
KLHL23
|
kelch-like 23 (Drosophila)
|
|
chr2_+_9264360
|
0.792
|
|
ASAP2
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2
|
|
chr7_+_26208532
|
0.782
|
|
|
|
|
chr6_-_13922525
|
0.780
|
NM_001031713
|
CCDC90A
|
coiled-coil domain containing 90A
|
|
chr12_-_37585602
|
0.778
|
NM_153634
|
CPNE8
|
copine VIII
|
|
chr12_+_109203412
|
0.778
|
NM_001135765
NM_001681
NM_170665
|
ATP2A2
|
ATPase, Ca++ transporting, cardiac muscle, slow twitch 2
|
|
chr1_+_213807321
|
0.770
|
NM_016121
|
KCTD3
|
potassium channel tetramerisation domain containing 3
|
|
chr17_+_32924049
|
0.769
|
NM_007026
|
DUSP14
|
dual specificity phosphatase 14
|
|
chr7_-_80386602
|
0.767
|
NM_006379
|
SEMA3C
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C
|
|
chr2_+_75039273
|
0.767
|
NM_019896
|
POLE4
|
polymerase (DNA-directed), epsilon 4 (p12 subunit)
|
|
chr7_-_80386258
|
0.761
|
|
SEMA3C
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C
|
|
chr2_+_149895274
|
0.758
|
NM_194317
|
LYPD6
|
LY6/PLAUR domain containing 6
|
|
chr1_+_36046373
|
0.757
|
NM_017629
|
EIF2C4
|
eukaryotic translation initiation factor 2C, 4
|
|
chr8_+_87595762
|
0.754
|
NM_003909
|
CPNE3
|
copine III
|
|
chr17_+_2443983
|
0.751
|
|
PAFAH1B1
|
platelet-activating factor acetylhydrolase 1b, regulatory subunit 1 (45kDa)
|
|
chr9_+_130491340
|
0.751
|
|
SET
|
SET nuclear oncogene
|
|
chr13_+_75021886
|
0.747
|
NM_006002
|
UCHL3
|
ubiquitin carboxyl-terminal esterase L3 (ubiquitin thiolesterase)
|
|
chr4_+_140594410
|
0.746
|
NM_031296
|
RAB33B
|
RAB33B, member RAS oncogene family
|
|
chr12_+_95112385
|
0.746
|
|
ELK3
|
ELK3, ETS-domain protein (SRF accessory protein 2)
|
|
chr2_+_28971007
|
0.741
|
NM_015131
|
WDR43
|
WD repeat domain 43
|
|
chr20_+_56899867
|
0.740
|
|
GNAS
|
GNAS complex locus
|
|
chr2_+_46779553
|
0.737
|
NM_014011
|
SOCS5
|
suppressor of cytokine signaling 5
|
|
chr12_+_6907646
|
0.736
|
NM_001940
|
ATN1
|
atrophin 1
|
|
chr16_+_30617962
|
0.736
|
NM_006662
|
SRCAP
|
Snf2-related CREBBP activator protein
|
|
chr4_+_99401543
|
0.736
|
NM_001100426
NM_001100427
NM_001100428
NM_001100429
NM_001100430
NM_021159
|
RAP1GDS1
|
RAP1, GTP-GDP dissociation stimulator 1
|
|
chr13_-_50315832
|
0.733
|
NM_198989
|
DLEU7
|
deleted in lymphocytic leukemia, 7
|
|
chr9_+_90193058
|
0.729
|
NM_006717
|
SPIN1
|
spindlin 1
|
|
chr8_+_87595782
|
0.719
|
|
CPNE3
|
copine III
|
|
chr8_+_28407613
|
0.715
|
NM_017412
|
FZD3
|
frizzled homolog 3 (Drosophila)
|
|
chr11_+_105453501
|
0.708
|
NM_015423
|
AASDHPPT
|
aminoadipate-semialdehyde dehydrogenase-phosphopantetheinyl transferase
|
|
chr1_-_58815753
|
0.707
|
NM_002353
|
TACSTD2
|
tumor-associated calcium signal transducer 2
|
|
chr12_+_75682016
|
0.706
|
|
ZDHHC17
|
zinc finger, DHHC-type containing 17
|
|
chr12_+_95112131
|
0.703
|
NM_005230
|
ELK3
|
ELK3, ETS-domain protein (SRF accessory protein 2)
|
|
chr8_+_22354276
|
0.701
|
|
PPP3CC
|
protein phosphatase 3, catalytic subunit, gamma isozyme
|
|
chr2_+_9264327
|
0.697
|
NM_001135191
NM_003887
|
ASAP2
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2
|
|
chr3_+_129253977
|
0.693
|
|
SEC61A1
|
Sec61 alpha 1 subunit (S. cerevisiae)
|
|
chr4_+_113286001
|
0.692
|
NM_152400
|
C4orf32
|
chromosome 4 open reading frame 32
|
|
chr4_-_110574363
|
0.689
|
|
1/2-SBSRNA4
|
lncRNA_BC009800
|
|
chr6_+_111911226
|
0.686
|
|
LOC643749
|
hypothetical LOC643749
|
|
chr15_-_29950188
|
0.679
|
|
OTUD7A
|
OTU domain containing 7A
|
|
chr13_-_40533072
|
0.678
|
|
ELF1
|
E74-like factor 1 (ets domain transcription factor)
|
|
chr2_+_206732859
|
0.677
|
|
EEF1B2
|
eukaryotic translation elongation factor 1 beta 2
|
|
chr3_+_73128629
|
0.675
|
|
PPP4R2
|
protein phosphatase 4, regulatory subunit 2
|
|
chr12_+_75681973
|
0.670
|
NM_015336
|
ZDHHC17
|
zinc finger, DHHC-type containing 17
|
|
chr13_+_97593711
|
0.669
|
|
FARP1
|
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived)
|
|
chr11_+_32069183
|
0.666
|
|
RCN1
|
reticulocalbin 1, EF-hand calcium binding domain
|
|
chr17_+_11864841
|
0.662
|
NM_003010
|
MAP2K4
|
mitogen-activated protein kinase kinase 4
|
|
chr3_+_185563872
|
0.661
|
NM_006232
|
POLR2H
|
polymerase (RNA) II (DNA directed) polypeptide H
|
|
chr1_-_62926533
|
0.659
|
NM_033407
|
DOCK7
|
dedicator of cytokinesis 7
|
|
chr16_+_56038837
|
0.658
|
NM_020312
|
COQ9
|
coenzyme Q9 homolog (S. cerevisiae)
|
|
chr16_+_83291107
|
0.658
|
|
USP10
|
ubiquitin specific peptidase 10
|
|
chr14_-_24588916
|
0.657
|
NM_014178
|
STXBP6
|
syntaxin binding protein 6 (amisyn)
|
|
chr17_+_57909979
|
0.656
|
NM_001112707
NM_006852
|
TLK2
|
tousled-like kinase 2
|
|
chr3_+_129253997
|
0.655
|
|
SEC61A1
|
Sec61 alpha 1 subunit (S. cerevisiae)
|
|
chr15_+_56850776
|
0.654
|
|
FAM63B
|
family with sequence similarity 63, member B
|
|
chr16_+_15397120
|
0.653
|
|
MPV17L
|
MPV17 mitochondrial membrane protein-like
|
|
chr3_+_134775247
|
0.650
|
|
CDV3
|
CDV3 homolog (mouse)
|
|
chr18_+_58341637
|
0.647
|
NM_017742
|
ZCCHC2
|
zinc finger, CCHC domain containing 2
|
|
chr10_-_104463947
|
0.646
|
NM_004311
|
ARL3
|
ADP-ribosylation factor-like 3
|
|
chr8_+_94998258
|
0.641
|
NM_018444
NM_001161780
NM_001161779
NM_001161781
|
PDP1
|
pyruvate dehyrogenase phosphatase catalytic subunit 1
|
|
chr11_+_8660913
|
0.635
|
|
RPL27A
|
ribosomal protein L27a
|
|
chr16_+_55523248
|
0.635
|
NM_001010989
NM_001010990
NM_014685
|
HERPUD1
|
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1
|
|
chr1_-_227828416
|
0.633
|
NM_001025247
NM_014409
|
TAF5L
|
TAF5-like RNA polymerase II, p300/CBP-associated factor (PCAF)-associated factor, 65kDa
|
|
chr2_+_138975819
|
0.633
|
NM_001001664
|
SPOPL
|
speckle-type POZ protein-like
|
|
chr20_+_6696744
|
0.631
|
NM_001200
|
BMP2
|
bone morphogenetic protein 2
|
|
chr3_+_73128667
|
0.631
|
|
PPP4R2
|
protein phosphatase 4, regulatory subunit 2
|
|
chr10_+_92970487
|
0.630
|
|
PCGF5
|
polycomb group ring finger 5
|
|
chr2_+_97142959
|
0.629
|
NM_001164315
|
ANKRD36
|
ankyrin repeat domain 36
|