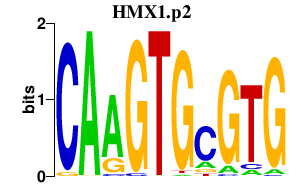
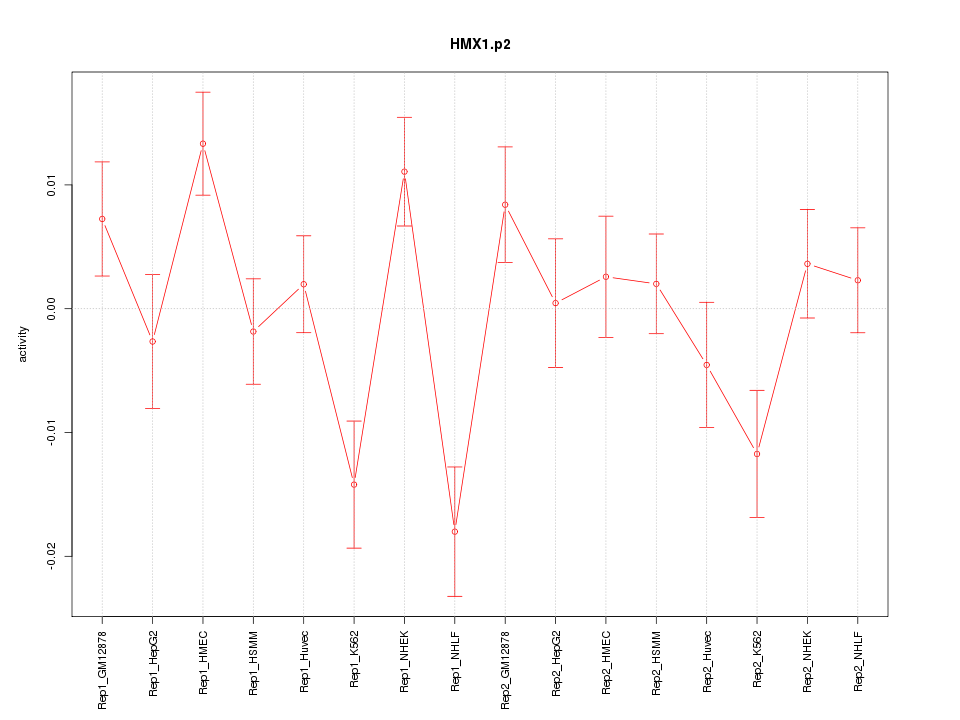
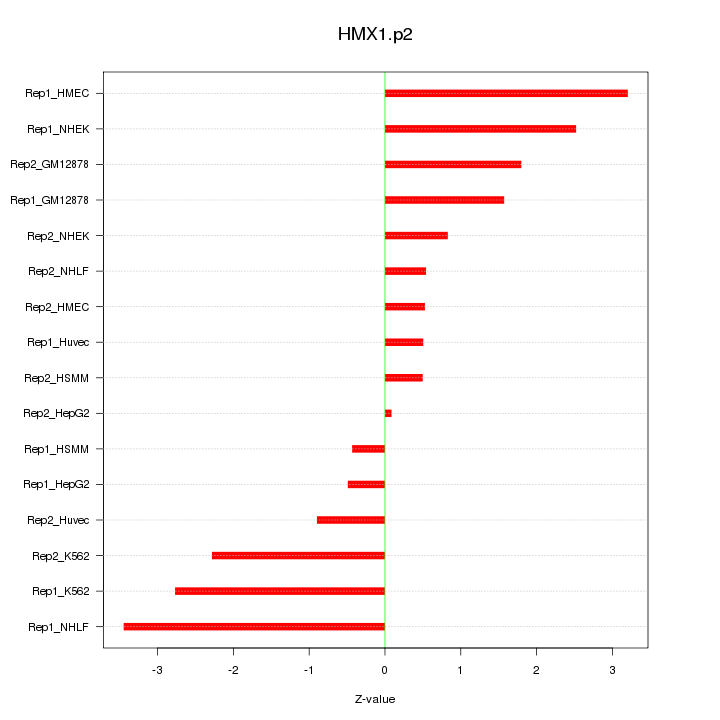
|
chr20_-_21442663
|
2.453
|
NM_002509
|
NKX2-2
|
NK2 homeobox 2
|
|
chr8_+_32525269
|
2.415
|
NM_001160002
NM_001160004
NM_001160005
NM_001160007
NM_001160008
NM_004495
NM_013956
NM_013957
NM_013958
NM_013960
NM_013964
|
NRG1
|
neuregulin 1
|
|
chr10_-_99521701
|
2.318
|
NM_003015
|
SFRP5
|
secreted frizzled-related protein 5
|
|
chr3_+_57969127
|
2.061
|
NM_001164317
NM_001164318
NM_001164319
NM_001457
|
FLNB
|
filamin B, beta
|
|
chr10_+_94811010
|
2.026
|
NM_183374
|
CYP26C1
|
cytochrome P450, family 26, subfamily C, polypeptide 1
|
|
chrX_+_48915127
|
1.946
|
NM_002668
|
PLP2
|
proteolipid protein 2 (colonic epithelium-enriched)
|
|
chr11_+_268569
|
1.830
|
NM_138329
|
NLRP6
|
NLR family, pyrin domain containing 6
|
|
chr19_+_590878
|
1.771
|
NM_020637
|
FGF22
|
fibroblast growth factor 22
|
|
chr19_+_43447078
|
1.641
|
|
SPINT2
|
serine peptidase inhibitor, Kunitz type, 2
|
|
chr15_-_75694674
|
1.597
|
|
LINGO1
|
leucine rich repeat and Ig domain containing 1
|
|
chr12_+_4253143
|
1.587
|
NM_001759
|
CCND2
|
cyclin D2
|
|
chr10_+_103882776
|
1.585
|
NM_015062
|
PPRC1
|
peroxisome proliferator-activated receptor gamma, coactivator-related 1
|
|
chr7_+_5609416
|
1.544
|
|
FSCN1
|
fascin homolog 1, actin-bundling protein (Strongylocentrotus purpuratus)
|
|
chr19_+_43446937
|
1.477
|
NM_001166103
NM_021102
|
SPINT2
|
serine peptidase inhibitor, Kunitz type, 2
|
|
chr19_+_43447262
|
1.433
|
|
SPINT2
|
serine peptidase inhibitor, Kunitz type, 2
|
|
chr20_+_2581177
|
1.377
|
NM_006392
|
NOP56
|
NOP56 ribonucleoprotein homolog (yeast)
|
|
chr22_-_35011703
|
1.363
|
|
MYH9
|
myosin, heavy chain 9, non-muscle
|
|
chr5_+_148186348
|
1.358
|
NM_000024
|
ADRB2
|
adrenergic, beta-2-, receptor, surface
|
|
chr20_-_538909
|
1.243
|
NM_004609
|
TCF15
|
transcription factor 15 (basic helix-loop-helix)
|
|
chr1_+_151917674
|
1.225
|
NM_000906
|
NPR1
|
natriuretic peptide receptor A/guanylate cyclase A (atrionatriuretic peptide receptor A)
|
|
chr16_+_87232501
|
1.186
|
NM_013278
|
IL17C
|
interleukin 17C
|
|
chrX_+_40367761
|
1.178
|
NM_001195522
|
LOC347411
|
brain protein 44-like protein 2
|
|
chr22_+_28032950
|
1.167
|
NM_006478
NM_152236
NM_152237
|
GAS2L1
|
growth arrest-specific 2 like 1
|
|
chr11_-_407324
|
1.150
|
NM_001135053
NM_021805
|
SIGIRR
|
single immunoglobulin and toll-interleukin 1 receptor (TIR) domain
|
|
chr17_+_1906262
|
1.135
|
NM_001098202
|
HIC1
|
hypermethylated in cancer 1
|
|
chr1_+_234372454
|
1.128
|
NM_003272
|
GPR137B
|
G protein-coupled receptor 137B
|
|
chr7_+_72486044
|
1.121
|
NM_003508
|
FZD9
|
frizzled homolog 9 (Drosophila)
|
|
chr11_+_66381451
|
1.118
|
NM_024036
|
LRFN4
|
leucine rich repeat and fibronectin type III domain containing 4
|
|
chr6_+_33697023
|
1.104
|
NM_002224
|
ITPR3
|
inositol 1,4,5-triphosphate receptor, type 3
|
|
chr10_+_83625049
|
1.099
|
NM_001010848
NM_001165972
|
NRG3
|
neuregulin 3
|
|
chr7_-_719268
|
1.094
|
NM_001164761
|
PRKAR1B
|
protein kinase, cAMP-dependent, regulatory, type I, beta
|
|
chr9_+_134843918
|
1.085
|
NM_001135031
NM_004188
|
GFI1B
|
growth factor independent 1B transcription repressor
|
|
chr16_-_85099941
|
1.073
|
|
LOC400550
|
hypothetical LOC400550
|
|
chr22_+_36384647
|
1.070
|
NM_020315
|
PDXP
|
pyridoxal (pyridoxine, vitamin B6) phosphatase
|
|
chr10_+_72102563
|
1.017
|
NM_080722
NM_139155
|
ADAMTS14
|
ADAM metallopeptidase with thrombospondin type 1 motif, 14
|
|
chr17_-_72045218
|
1.007
|
NM_134268
|
CYGB
|
cytoglobin
|
|
chr12_+_93066370
|
0.997
|
NM_005761
|
PLXNC1
|
plexin C1
|
|
chr2_+_242146791
|
0.989
|
NM_032515
|
BOK
|
BCL2-related ovarian killer
|
|
chr19_-_54857003
|
0.985
|
|
IRF3
|
interferon regulatory factor 3
|
|
chrX_+_13497614
|
0.976
|
NM_001167890
NM_015507
|
EGFL6
|
EGF-like-domain, multiple 6
|
|
chr17_-_76064842
|
0.971
|
NM_002522
|
NPTX1
|
neuronal pentraxin I
|
|
chr14_+_22845851
|
0.968
|
NM_004050
|
BCL2L2-PABPN1
BCL2L2
|
BCL2L2-PABPN1 read-through transcript
BCL2-like 2
|
|
chr2_-_233060774
|
0.966
|
NM_004826
|
ECEL1
|
endothelin converting enzyme-like 1
|
|
chr7_-_19151509
|
0.964
|
NM_152898
|
FERD3L
|
Fer3-like (Drosophila)
|
|
chr2_-_241408296
|
0.959
|
NM_004321
|
KIF1A
|
kinesin family member 1A
|
|
chr1_+_1557419
|
0.943
|
NM_006983
|
MMP23B
|
matrix metallopeptidase 23B
|
|
chr16_+_3002457
|
0.926
|
NM_020982
|
CLDN9
|
claudin 9
|
|
chr14_+_41146513
|
0.906
|
NM_152447
|
LRFN5
|
leucine rich repeat and fibronectin type III domain containing 5
|
|
chr12_+_4300607
|
0.882
|
NM_020375
|
C12orf5
|
chromosome 12 open reading frame 5
|
|
chr14_-_88328848
|
0.842
|
NM_183387
|
EML5
|
echinoderm microtubule associated protein like 5
|
|
chr2_+_218833982
|
0.834
|
NM_001077191
NM_001077194
NM_170699
|
GPBAR1
|
G protein-coupled bile acid receptor 1
|
|
chr8_+_32623792
|
0.831
|
NM_013959
|
NRG1
|
neuregulin 1
|
|
chr12_-_47679459
|
0.816
|
|
DDN
|
dendrin
|
|
chr16_-_1763036
|
0.809
|
NM_023936
|
MRPS34
|
mitochondrial ribosomal protein S34
|
|
chr11_+_66381077
|
0.799
|
|
LRFN4
|
leucine rich repeat and fibronectin type III domain containing 4
|
|
chr19_+_58733144
|
0.796
|
NM_001079906
|
ZNF331
|
zinc finger protein 331
|
|
chr4_-_2390050
|
0.793
|
NM_001172656
NM_001172657
NM_001172658
NM_020972
|
ZFYVE28
|
zinc finger, FYVE domain containing 28
|
|
chr21_-_26345294
|
0.791
|
|
|
|
|
chr16_-_65751263
|
0.780
|
|
TRADD
|
TNFRSF1A-associated via death domain
|
|
chr2_+_109729176
|
0.772
|
NM_023016
|
ANKRD57
|
ankyrin repeat domain 57
|
|
chr17_+_71229191
|
0.769
|
|
ITGB4
|
integrin, beta 4
|
|
chr16_-_65751305
|
0.761
|
NM_003789
|
TRADD
|
TNFRSF1A-associated via death domain
|
|
chr3_-_129777618
|
0.761
|
NM_007354
|
C3orf27
|
chromosome 3 open reading frame 27
|
|
chr17_-_77635703
|
0.754
|
|
FASN
|
fatty acid synthase
|
|
chr14_-_104705753
|
0.753
|
|
JAG2
|
jagged 2
|
|
chr14_+_101097440
|
0.752
|
NM_001362
|
DIO3
|
deiodinase, iodothyronine, type III
|
|
chr9_-_26882713
|
0.749
|
NM_001167575
NM_024828
|
C9orf82
|
chromosome 9 open reading frame 82
|
|
chr14_+_73775927
|
0.746
|
NM_182894
|
VSX2
|
visual system homeobox 2
|
|
chr10_+_126140330
|
0.743
|
NM_001167880
NM_022126
|
LHPP
|
phospholysine phosphohistidine inorganic pyrophosphate phosphatase
|
|
chr17_-_15185623
|
0.737
|
NM_031898
|
TEKT3
|
tektin 3
|
|
chr4_-_142273881
|
0.732
|
NM_020724
|
RNF150
|
ring finger protein 150
|
|
chr14_+_100270222
|
0.730
|
|
DLK1
|
delta-like 1 homolog (Drosophila)
|
|
chr19_+_2240996
|
0.715
|
|
|
|
|
chr11_-_45264193
|
0.711
|
NM_020826
|
SYT13
|
synaptotagmin XIII
|
|
chr4_+_2390469
|
0.710
|
NM_001193282
|
LOC402160
|
hypothetical protein LOC402160
|
|
chr17_+_21220260
|
0.709
|
NM_021012
|
KCNJ12
|
potassium inwardly-rectifying channel, subfamily J, member 12
|
|
chr8_+_31616809
|
0.707
|
NM_013962
|
NRG1
|
neuregulin 1
|
|
chr18_-_45975163
|
0.702
|
NM_001080467
|
MYO5B
|
myosin VB
|
|
chr4_+_75529716
|
0.700
|
NM_001657
|
AREG
|
amphiregulin
|
|
chr17_+_19255083
|
0.696
|
NM_007148
|
RNF112
|
ring finger protein 112
|
|
chr1_-_215377718
|
0.693
|
NM_001134285
|
ESRRG
|
estrogen-related receptor gamma
|
|
chrX_-_100332933
|
0.691
|
|
YWHAQP8
|
YWHAQ pseudogene 8
|
|
chr7_-_719102
|
0.687
|
NM_001164762
|
PRKAR1B
|
protein kinase, cAMP-dependent, regulatory, type I, beta
|
|
chr18_+_48120539
|
0.685
|
NM_005215
|
DCC
|
deleted in colorectal carcinoma
|
|
chr1_+_39229474
|
0.675
|
NM_001136275
NM_024595
|
AKIRIN1
|
akirin 1
|
|
chr18_-_55091596
|
0.675
|
NM_013435
|
RAX
|
retina and anterior neural fold homeobox
|
|
chr22_+_35777561
|
0.672
|
NM_024681
|
KCTD17
|
potassium channel tetramerisation domain containing 17
|
|
chr19_+_4445291
|
0.665
|
|
HDGFRP2
|
hepatoma-derived growth factor-related protein 2
|
|
chr3_+_127743868
|
0.658
|
|
CHST13
|
carbohydrate (chondroitin 4) sulfotransferase 13
|
|
chr8_+_120497732
|
0.651
|
NM_002514
|
NOV
|
nephroblastoma overexpressed gene
|
|
chr3_+_122109716
|
0.650
|
NM_014980
|
STXBP5L
|
syntaxin binding protein 5-like
|
|
chrX_-_131918905
|
0.648
|
|
HS6ST2
|
heparan sulfate 6-O-sulfotransferase 2
|
|
chr20_+_20296744
|
0.648
|
NM_002196
|
INSM1
|
insulinoma-associated 1
|
|
chr2_+_171381317
|
0.645
|
NM_000817
NM_013445
|
GAD1
|
glutamate decarboxylase 1 (brain, 67kDa)
|
|
chr15_-_75693309
|
0.645
|
|
LINGO1
|
leucine rich repeat and Ig domain containing 1
|
|
chr6_+_41714171
|
0.644
|
NM_005586
|
MDFI
|
MyoD family inhibitor
|
|
chr17_-_3651236
|
0.643
|
NM_002208
|
ITGAE
|
integrin, alpha E (antigen CD103, human mucosal lymphocyte antigen 1; alpha polypeptide)
|
|
chr1_+_180836050
|
0.641
|
|
|
|
|
chr19_-_10202947
|
0.623
|
NM_004230
|
S1PR2
|
sphingosine-1-phosphate receptor 2
|
|
chr3_+_127118130
|
0.621
|
|
LOC100125556
|
family with sequence similarity 86, member A pseudogene
|
|
chr22_-_49063088
|
0.620
|
|
PLXNB2
|
plexin B2
|
|
chr12_-_42231990
|
0.612
|
NM_025003
|
ADAMTS20
|
ADAM metallopeptidase with thrombospondin type 1 motif, 20
|
|
chr19_+_627346
|
0.609
|
NM_005860
|
FSTL3
|
follistatin-like 3 (secreted glycoprotein)
|
|
chr2_-_122210873
|
0.607
|
NM_032390
|
MKI67IP
|
MKI67 (FHA domain) interacting nucleolar phosphoprotein
|
|
chr12_-_47651812
|
0.603
|
NM_003394
|
WNT10B
|
wingless-type MMTV integration site family, member 10B
|
|
chr4_-_109307325
|
0.602
|
NM_001166119
|
LEF1
|
lymphoid enhancer-binding factor 1
|
|
chr3_-_193928038
|
0.599
|
NM_004113
|
FGF12
|
fibroblast growth factor 12
|
|
chr4_-_21559471
|
0.598
|
NM_147182
|
KCNIP4
|
Kv channel interacting protein 4
|
|
chr20_+_2974659
|
0.597
|
NM_030811
|
MRPS26
|
mitochondrial ribosomal protein S26
|
|
chr10_-_135000464
|
0.593
|
NM_015722
|
CALY
|
calcyon neuron-specific vesicular protein
|
|
chr5_+_170779664
|
0.593
|
|
FGF18
|
fibroblast growth factor 18
|
|
chr15_-_38953628
|
0.591
|
NM_133639
|
RHOV
|
ras homolog gene family, member V
|
|
chr20_-_26137755
|
0.588
|
|
LOC284801
|
hypothetical protein LOC284801
|
|
chr7_+_30917939
|
0.583
|
NM_198098
|
AQP1
|
aquaporin 1 (Colton blood group)
|
|
chr20_+_21231921
|
0.575
|
NM_012255
|
XRN2
|
5'-3' exoribonuclease 2
|
|
chr16_+_1763185
|
0.572
|
NM_001010865
|
EME2
|
essential meiotic endonuclease 1 homolog 2 (S. pombe)
|
|
chr11_+_63758550
|
0.571
|
NM_003377
|
VEGFB
|
vascular endothelial growth factor B
|
|
chr16_+_56259657
|
0.570
|
NM_170776
|
GPR97
|
G protein-coupled receptor 97
|
|
chr3_-_57088332
|
0.568
|
NM_001128615
|
ARHGEF3
|
Rho guanine nucleotide exchange factor (GEF) 3
|
|
chr7_-_97719372
|
0.566
|
NM_015395
|
TECPR1
|
tectonin beta-propeller repeat containing 1
|
|
chr7_-_100009205
|
0.566
|
NM_001168682
|
SAP25
|
sin3A-binding protein 25
|
|
chr4_+_2031036
|
0.565
|
NM_178557
|
NAT8L
|
N-acetyltransferase 8-like (GCN5-related, putative)
|
|
chr16_+_12902955
|
0.564
|
NM_001145204
NM_001145205
|
SHISA9
|
shisa homolog 9 (Xenopus laevis)
|
|
chr10_-_98470164
|
0.560
|
NM_152309
|
PIK3AP1
|
phosphoinositide-3-kinase adaptor protein 1
|
|
chr8_-_26780665
|
0.556
|
|
ADRA1A
|
adrenergic, alpha-1A-, receptor
|
|
chr6_+_36754467
|
0.556
|
|
CDKN1A
|
cyclin-dependent kinase inhibitor 1A (p21, Cip1)
|
|
chr17_-_9635325
|
0.548
|
NM_001105571
|
DHRS7C
|
dehydrogenase/reductase (SDR family) member 7C
|
|
chr20_+_30103642
|
0.546
|
NM_001172129
NM_001172130
NM_001172131
NM_001172132
NM_001172133
NM_002110
|
HCK
|
hemopoietic cell kinase
|
|
chr16_+_65754788
|
0.546
|
NM_001040667
NM_001538
|
HSF4
|
heat shock transcription factor 4
|
|
chr19_+_47056162
|
0.541
|
|
RPS19
|
ribosomal protein S19
|
|
chr2_+_176677479
|
0.541
|
|
|
|
|
chr20_+_60311469
|
0.535
|
|
ADRM1
|
adhesion regulating molecule 1
|
|
chr12_-_47679176
|
0.535
|
NM_015086
|
DDN
|
dendrin
|
|
chr16_+_2954217
|
0.535
|
NM_024507
NM_172229
|
KREMEN2
|
kringle containing transmembrane protein 2
|
|
chr11_+_66547391
|
0.534
|
NM_177963
|
SYT12
|
synaptotagmin XII
|
|
chrX_+_46656543
|
0.532
|
NM_001077445
NM_014735
|
PHF16
|
PHD finger protein 16
|
|
chr13_-_75009941
|
0.532
|
NM_203495
NM_203497
|
COMMD6
|
COMM domain containing 6
|
|
chr12_-_15265490
|
0.532
|
NM_001190726
NM_032918
|
RERG
|
RAS-like, estrogen-regulated, growth inhibitor
|
|
chr9_-_138277467
|
0.526
|
NM_181701
|
QSOX2
|
quiescin Q6 sulfhydryl oxidase 2
|
|
chr2_+_242289983
|
0.526
|
NM_032329
|
ING5
|
inhibitor of growth family, member 5
|
|
chr9_-_10602722
|
0.525
|
NM_002839
|
PTPRD
|
protein tyrosine phosphatase, receptor type, D
|
|
chr14_+_61653827
|
0.524
|
|
FLJ43390
|
hypothetical LOC646113
|
|
chr17_+_58908152
|
0.521
|
NM_000789
|
ACE
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 1
|
|
chr19_+_47056102
|
0.520
|
|
RPS19
|
ribosomal protein S19
|
|
chr9_-_34554800
|
0.520
|
|
CNTFR
|
ciliary neurotrophic factor receptor
|
|
chr20_+_2224612
|
0.518
|
NM_003245
|
TGM3
|
transglutaminase 3 (E polypeptide, protein-glutamine-gamma-glutamyltransferase)
|
|
chrX_-_111809929
|
0.518
|
NM_178175
|
LHFPL1
|
lipoma HMGIC fusion partner-like 1
|
|
chr5_+_156625629
|
0.518
|
NM_001037333
NM_001037332
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2
|
|
chrX_+_101853912
|
0.516
|
NM_001004051
NM_001184874
NM_001184875
NM_001184876
NM_138437
|
GPRASP2
|
G protein-coupled receptor associated sorting protein 2
|
|
chr14_+_20568184
|
0.515
|
NM_173846
|
TPPP2
|
tubulin polymerization-promoting protein family member 2
|
|
chr3_+_50291521
|
0.514
|
NM_153215
|
C3orf45
|
chromosome 3 open reading frame 45
|
|
chr17_+_9489578
|
0.513
|
NM_153210
|
USP43
|
ubiquitin specific peptidase 43
|
|
chr9_-_23811477
|
0.512
|
NM_001171197
|
ELAVL2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr1_-_45081190
|
0.511
|
NM_001166292
NM_003738
|
PTCH2
|
patched 2
|
|
chr10_+_106390848
|
0.511
|
NM_014978
|
SORCS3
|
sortilin-related VPS10 domain containing receptor 3
|
|
chr7_+_50314801
|
0.510
|
NM_006060
|
IKZF1
|
IKAROS family zinc finger 1 (Ikaros)
|
|
chr15_+_62230962
|
0.509
|
NM_024798
|
SNX22
|
sorting nexin 22
|
|
chr2_-_27469947
|
0.509
|
|
FTH1P3
|
ferritin, heavy polypeptide 1 pseudogene 3
|
|
chrX_-_152863251
|
0.509
|
NM_002910
|
RENBP
|
renin binding protein
|
|
chr19_+_40474867
|
0.508
|
NM_002361
NM_080600
|
MAG
|
myelin associated glycoprotein
|
|
chr5_-_172594691
|
0.507
|
NM_001166175
NM_001166176
NM_004387
|
NKX2-5
|
NK2 transcription factor related, locus 5 (Drosophila)
|
|
chr20_+_61655934
|
0.507
|
|
|
|
|
chr17_+_71229110
|
0.506
|
NM_000213
NM_001005731
|
ITGB4
|
integrin, beta 4
|
|
chr10_+_72826656
|
0.506
|
NM_001171930
NM_001171931
NM_001171932
NM_022124
NM_052836
|
CDH23
|
cadherin-related 23
|
|
chr12_-_47604930
|
0.506
|
NM_001143781
|
FKBP11
|
FK506 binding protein 11, 19 kDa
|
|
chr19_+_40299058
|
0.504
|
|
FXYD3
|
FXYD domain containing ion transport regulator 3
|
|
chr4_+_81325447
|
0.503
|
NM_020226
|
PRDM8
|
PR domain containing 8
|
|
chr9_+_966963
|
0.501
|
NM_021240
|
DMRT3
|
doublesex and mab-3 related transcription factor 3
|
|
chr21_-_38954433
|
0.499
|
|
ERG
|
v-ets erythroblastosis virus E26 oncogene homolog (avian)
|
|
chr11_+_1367665
|
0.498
|
NM_003957
|
BRSK2
|
BR serine/threonine kinase 2
|
|
chr22_+_45537181
|
0.496
|
NM_014346
|
TBC1D22A
|
TBC1 domain family, member 22A
|
|
chr3_-_52839727
|
0.495
|
NM_001166449
NM_002218
|
ITIH4
|
inter-alpha (globulin) inhibitor H4 (plasma Kallikrein-sensitive glycoprotein)
|
|
chr15_+_87147667
|
0.494
|
NM_001135
NM_013227
|
ACAN
|
aggrecan
|
|
chr14_-_100104014
|
0.493
|
NM_001159531
|
BEGAIN
|
brain-enriched guanylate kinase-associated homolog (rat)
|
|
chr16_-_87405817
|
0.492
|
NM_000485
NM_001030018
|
APRT
|
adenine phosphoribosyltransferase
|
|
chr2_-_219566364
|
0.492
|
NM_005209
NM_057093
NM_057094
|
CRYBA2
|
crystallin, beta A2
|
|
chr11_+_527521
|
0.492
|
NM_198075
|
LRRC56
|
leucine rich repeat containing 56
|
|
chrX_+_46318135
|
0.491
|
NM_019886
|
CHST7
|
carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 7
|
|
chr1_-_194844061
|
0.489
|
NM_198503
|
KCNT2
|
potassium channel, subfamily T, member 2
|
|
chr1_-_194844299
|
0.489
|
|
KCNT2
|
potassium channel, subfamily T, member 2
|
|
chr1_+_35020425
|
0.489
|
NM_001005752
|
GJB3
|
gap junction protein, beta 3, 31kDa
|
|
chr19_+_62523636
|
0.488
|
NM_213598
|
ZNF543
|
zinc finger protein 543
|
|
chr9_-_135029096
|
0.487
|
NM_021996
|
GBGT1
RALGDS
|
globoside alpha-1,3-N-acetylgalactosaminyltransferase 1
ral guanine nucleotide dissociation stimulator
|
|
chr22_+_28446288
|
0.486
|
NM_182527
|
CABP7
|
calcium binding protein 7
|
|
chr1_+_154478574
|
0.483
|
NM_199173
|
BGLAP
|
bone gamma-carboxyglutamate (gla) protein
|
|
chrX_-_34585287
|
0.483
|
NM_031442
|
TMEM47
|
transmembrane protein 47
|
|
chr14_+_28306633
|
0.481
|
|
FOXG1
|
forkhead box G1
|
|
chr8_+_121206478
|
0.481
|
NM_021110
|
COL14A1
|
collagen, type XIV, alpha 1
|
|
chr19_+_52215982
|
0.479
|
NM_002517
|
NPAS1
|
neuronal PAS domain protein 1
|
|
chr11_+_69165065
|
0.479
|
|
CCND1
|
cyclin D1
|
|
chr5_-_88004891
|
0.477
|
|
LOC645323
|
hypothetical LOC645323
|
|
chr22_-_29272668
|
0.475
|
NM_001193336
|
LOC730005
|
SEC14-like (S. cerevisiae)
|
|
chr22_+_49128583
|
0.473
|
NM_014678
|
PPP6R2
|
protein phosphatase 6, regulatory subunit 2
|
|
chr9_+_139202874
|
0.472
|
NM_003731
|
SSNA1
|
Sjogren syndrome nuclear autoantigen 1
|
|
chr17_-_37275142
|
0.465
|
NM_018143
|
KLHL11
|
kelch-like 11 (Drosophila)
|
|
chr16_-_47872973
|
0.464
|
|
CBLN1
|
cerebellin 1 precursor
|
|
chr15_+_81567327
|
0.461
|
NM_001144903
NM_023003
|
TM6SF1
|
transmembrane 6 superfamily member 1
|
|
chrX_-_139414890
|
0.460
|
NM_005634
|
SOX3
|
SRY (sex determining region Y)-box 3
|
|
chr16_-_47872880
|
0.460
|
|
|
|
|
chr1_-_2554288
|
0.458
|
NM_033467
|
MMEL1
|
membrane metallo-endopeptidase-like 1
|