|
chrX_-_102870207
|
0.888
|
NM_001024452
NM_001172285
|
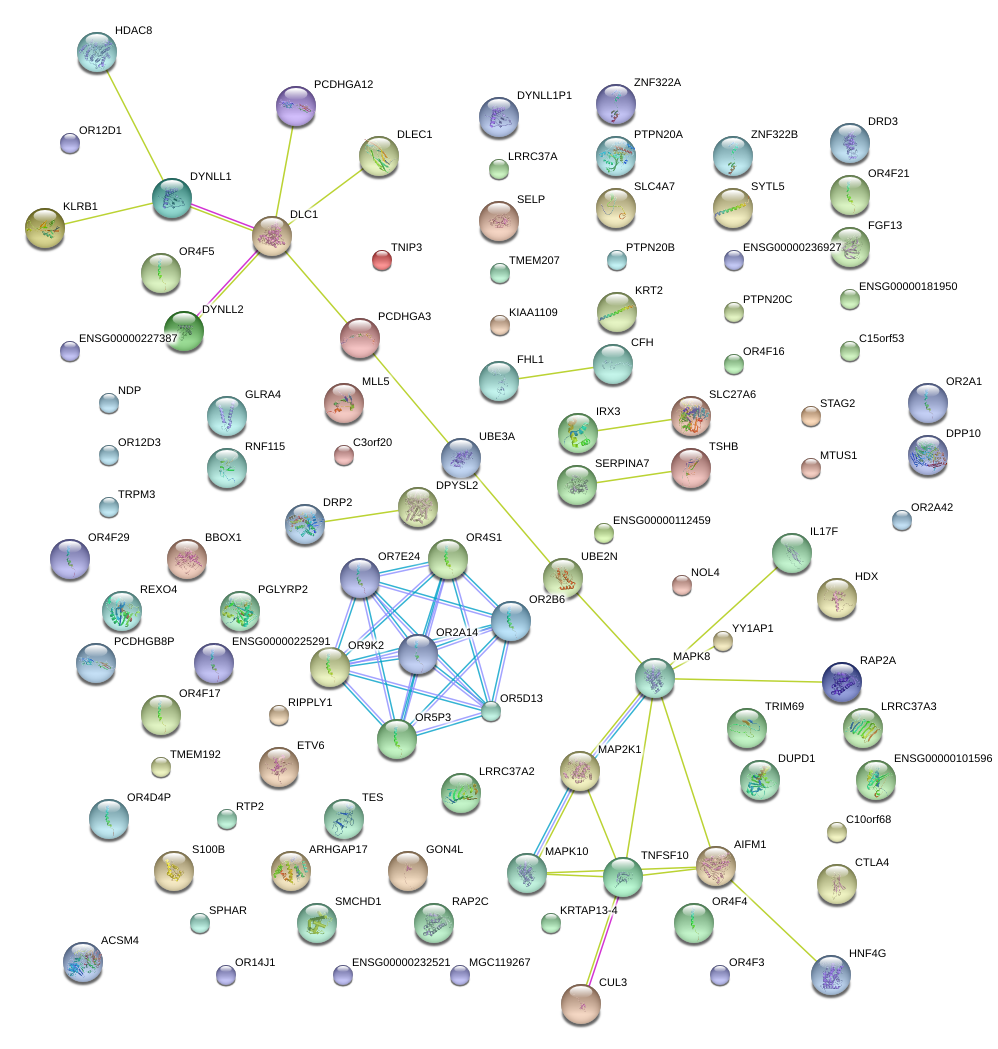
GLRA4
|
glycine receptor, alpha 4
|
|
chrX_-_105169370
|
0.872
|
NM_000354
|
SERPINA7
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 7
|
|
chr7_+_115650240
|
0.737
|
NM_152829
|
TES
|
testis derived transcript (3 LIM domains)
|
|
chr15_+_64481270
|
0.681
|
|
MAP2K1
|
mitogen-activated protein kinase kinase 1
|
|
chr12_+_7348194
|
0.663
|
NM_001080454
|
ACSM4
|
acyl-CoA synthetase medium-chain family member 4
|
|
chr4_+_123311207
|
0.655
|
NM_015312
|
KIAA1109
|
KIAA1109
|
|
chr1_-_611896
|
0.619
|
NM_001005224
NM_001005221
NM_001005277
|
OR4F3
OR4F29
OR4F16
|
olfactory receptor, family 4, subfamily F, member 3
olfactory receptor, family 4, subfamily F, member 29
olfactory receptor, family 4, subfamily F, member 16
|
|
chr21_+_30724442
|
0.548
|
NM_181600
|
KRTAP13-4
|
keratin associated protein 13-4
|
|
chr18_+_2645885
|
0.496
|
NM_015295
|
SMCHD1
|
structural maintenance of chromosomes flexible hinge domain containing 1
|
|
chr10_-_76488277
|
0.481
|
NM_001003892
|
DUPD1
|
dual specificity phosphatase and pro isomerase domain containing 1
|
|
chr18_-_29882555
|
0.459
|
NM_001198549
|
NOL4
|
nucleolar protein 4
|
|
chr2_+_204440753
|
0.450
|
NM_001037631
NM_005214
|
CTLA4
|
cytotoxic T-lymphocyte-associated protein 4
|
|
chr15_-_23204887
|
0.435
|
NM_000462
|
UBE3A
|
ubiquitin protein ligase E3A
|
|
chr1_-_154041393
|
0.434
|
|
GON4L
|
gon-4-like (C. elegans)
|
|
chrX_-_131181141
|
0.431
|
|
RAP2C
|
RAP2C, member of RAS oncogene family
|
|
chrX_+_135106578
|
0.412
|
NM_001159701
NM_001159699
|
FHL1
|
four and a half LIM domains 1
|
|
chrX_-_130792297
|
0.387
|
|
LOC286467
|
hypothetical LOC286467
|
|
chr3_-_191650291
|
0.386
|
NM_207316
|
TMEM207
|
transmembrane protein 207
|
|
chr6_-_29451042
|
0.381
|
NM_030959
|
OR12D3
|
olfactory receptor, family 12, subfamily D, member 3
|
|
chr1_+_115373937
|
0.368
|
NM_000549
|
TSHB
|
thyroid stimulating hormone, beta
|
|
chrX_+_37750778
|
0.356
|
NM_001163335
NM_138780
|
SYTL5
|
synaptotagmin-like 5
|
|
chr17_+_53587492
|
0.344
|
NM_012374
|
OR4D1
|
olfactory receptor, family 4, subfamily D, member 1
|
|
chr11_+_27019084
|
0.343
|
NM_003986
|
BBOX1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1
|
|
chr6_-_52217185
|
0.337
|
NM_052872
|
IL17F
|
interleukin 17F
|
|
chrX_-_83644054
|
0.304
|
NM_001177478
NM_001177479
NM_144657
|
HDX
|
highly divergent homeobox
|
|
chr7_+_104468614
|
0.290
|
|
MLL5
|
myeloid/lymphoid or mixed-lineage leukemia 5 (trithorax homolog, Drosophila)
|
|
chrX_-_43717864
|
0.285
|
NM_000266
|
NDP
|
Norrie disease (pseudoglioma)
|
|
chr10_+_32896656
|
0.276
|
NM_024688
|
C10orf68
|
chromosome 10 open reading frame 68
|
|
chr4_-_166253453
|
0.274
|
NM_001100389
|
TMEM192
|
transmembrane protein 192
|
|
chr2_+_114916368
|
0.264
|
NM_020868
|
DPP10
|
dipeptidyl-peptidase 10 (non-functional)
|
|
chr21_-_46849413
|
0.263
|
NM_006272
|
S100B
|
S100 calcium binding protein B
|
|
chr12_-_51332214
|
0.261
|
NM_000423
|
KRT2
|
keratin 2
|
|
chrX_-_71709335
|
0.238
|
|
HDAC8
|
histone deacetylase 8
|
|
chr11_+_55297489
|
0.237
|
NM_001001967
|
OR5D13
|
olfactory receptor, family 5, subfamily D, member 13
|
|
chrX_-_137777382
|
0.237
|
NM_001139502
|
FGF13
|
fibroblast growth factor 13
|
|
chr3_-_173723754
|
0.226
|
NM_001190943
NM_001190942
NM_003810
|
TNFSF10
|
tumor necrosis factor (ligand) superfamily, member 10
|
|
chr12_+_11694172
|
0.219
|
|
ETV6
|
ets variant 6
|
|
chr3_-_115380539
|
0.216
|
NM_000796
NM_033663
|
DRD3
|
dopamine receptor D3
|
|
chr12_+_53809731
|
0.215
|
NM_001005243
|
OR9K2
|
olfactory receptor, family 9, subfamily K, member 2
|
|
chrX_+_100361588
|
0.210
|
NM_001171184
NM_001939
|
DRP2
|
dystrophin related protein 2
|
|
chr8_-_17599438
|
0.209
|
NM_020749
|
MTUS1
|
microtubule associated tumor suppressor 1
|
|
chr10_-_46039941
|
0.207
|
NM_001042357
NM_001042389
|
PTPN20B
PTPN20A
|
protein tyrosine phosphatase, non-receptor type 20B
protein tyrosine phosphatase, non-receptor type 20A
|
|
chr7_+_143457083
|
0.202
|
NM_001001659
|
OR2A14
|
olfactory receptor, family 2, subfamily A, member 14
|
|
chr15_+_42815851
|
0.198
|
NM_080745
NM_182985
|
TRIM69
|
tripartite motif containing 69
|
|
chrX_-_129099695
|
0.196
|
NM_001130846
|
AIFM1
|
apoptosis-inducing factor, mitochondrion-associated, 1
|
|
chr16_-_52877868
|
0.194
|
NM_024336
|
IRX3
|
iroquois homeobox 3
|
|
chr1_-_167865973
|
0.187
|
NM_003005
|
SELP
|
selectin P (granule membrane protein 140kDa, antigen CD62)
|
|
chr4_-_122357104
|
0.187
|
NM_001128843
|
TNIP3
|
TNFAIP3 interacting protein 3
|
|
chr19_+_61624
|
0.187
|
NM_001005240
|
OR4F4
OR4F17
|
olfactory receptor, family 4, subfamily F, member 4
olfactory receptor, family 4, subfamily F, member 17
|
|
chr3_-_27473248
|
0.187
|
NM_003615
|
SLC4A7
|
solute carrier family 4, sodium bicarbonate cotransporter, member 7
|
|
chr9_-_72926333
|
0.184
|
NM_001007471
|
TRPM3
|
transient receptor potential cation channel, subfamily M, member 3
|
|
chr4_-_166253374
|
0.181
|
|
TMEM192
|
transmembrane protein 192
|
|
chr7_-_143560850
|
0.180
|
NM_001005287
NM_001001802
|
OR2A1
OR2A42
|
olfactory receptor, family 2, subfamily A, member 1
olfactory receptor, family 2, subfamily A, member 42
|
|
chr1_+_58899
|
0.177
|
NM_001005484
|
OR4F4
OR4F5
|
olfactory receptor, family 4, subfamily F, member 4
olfactory receptor, family 4, subfamily F, member 5
|
|
chr19_-_15451203
|
0.167
|
NM_052890
|
PGLYRP2
|
peptidoglycan recognition protein 2
|
|
chr11_+_48284350
|
0.166
|
NM_001004725
|
OR4S1
|
olfactory receptor, family 4, subfamily S, member 1
|
|
chr7_+_143646150
|
0.165
|
NM_001005287
NM_001001802
|
OR2A1
OR2A42
|
olfactory receptor, family 2, subfamily A, member 1
olfactory receptor, family 2, subfamily A, member 42
|
|
chr19_+_9222605
|
0.164
|
NM_001079935
|
OR7E24
|
olfactory receptor, family 7, subfamily E, member 24
|
|
chrX_+_123062143
|
0.159
|
|
STAG2
|
stromal antigen 2
|
|
chr5_+_140780720
|
0.156
|
NM_018914
NM_032091
NM_032092
|
PCDHGA11
|
protocadherin gamma subfamily A, 11
|
|
chr15_+_36776090
|
0.151
|
NM_207444
|
C15orf53
|
chromosome 15 open reading frame 53
|
|
chr1_+_227506751
|
0.143
|
NM_006542
|
SPHAR
|
S-phase response (cyclin related)
|
|
chr1_+_357502
|
0.143
|
NM_001005224
NM_001005221
NM_001005277
|
OR4F3
OR4F29
OR4F16
|
olfactory receptor, family 4, subfamily F, member 3
olfactory receptor, family 4, subfamily F, member 29
olfactory receptor, family 4, subfamily F, member 16
|
|
chr4_-_87247829
|
0.141
|
NM_138981
|
MAPK10
|
mitogen-activated protein kinase 10
|
|
chr3_-_188903038
|
0.140
|
NM_001004312
|
RTP2
|
receptor (chemosensory) transporter protein 2
|
|
chr8_+_76614757
|
0.140
|
NM_004133
|
HNF4G
|
hepatocyte nuclear factor 4, gamma
|
|
chr3_+_14691609
|
0.139
|
NM_001184957
NM_001184958
NM_032137
|
C3orf20
|
chromosome 3 open reading frame 20
|
|
chr5_+_128328685
|
0.139
|
|
SLC27A6
|
solute carrier family 27 (fatty acid transporter), member 6
|
|
chr6_+_29382381
|
0.135
|
NM_030946
|
OR14J1
|
olfactory receptor, family 14, subfamily J, member 1
|
|
chr1_+_144374513
|
0.134
|
|
RNF115
|
ring finger protein 115
|
|
chrX_-_106033202
|
0.134
|
NM_001171706
NM_138382
|
RIPPLY1
|
ripply1 homolog (zebrafish)
|
|
chr9_-_99001551
|
0.132
|
NM_199005
|
ZNF322B
ZNF322A
|
zinc finger protein 322B
zinc finger protein 322A
|
|
chr11_-_7804094
|
0.129
|
NM_153445
|
OR5P3
|
olfactory receptor, family 5, subfamily P, member 3
|
|
chr2_-_225078931
|
0.126
|
|
CUL3
|
cullin 3
|
|
chr17_+_41728273
|
0.125
|
NM_014834
|
LRRC37A
|
leucine rich repeat containing 37A
|
|
chr16_-_24850163
|
0.124
|
|
ARHGAP17
|
Rho GTPase activating protein 17
|
|
chr8_-_13018123
|
0.124
|
NM_001164271
|
DLC1
|
deleted in liver cancer 1
|
|
chr13_+_78068246
|
0.124
|
|
LOC780529
|
hypothetical LOC780529
|
|
chr17_+_17548888
|
0.123
|
|
|
|
|
chr14_+_19285426
|
0.123
|
NM_172194
|
OR4Q3
|
olfactory receptor, family 4, subfamily Q, member 3
|
|
chr3_+_58459131
|
0.123
|
NM_153331
|
KCTD6
|
potassium channel tetramerisation domain containing 6
|
|
chr10_+_98730977
|
0.122
|
NM_015652
|
C10orf12
|
chromosome 10 open reading frame 12
|
|
chr11_+_68124400
|
0.122
|
|
PPP6R3
|
protein phosphatase 6, regulatory subunit 3
|
|
chr14_+_19473606
|
0.122
|
NM_001004063
|
OR4K1
|
olfactory receptor, family 4, subfamily K, member 1
|
|
chr5_-_160044844
|
0.121
|
|
ATP10B
|
ATPase, class V, type 10B
|
|
chr22_+_23921072
|
0.121
|
|
KIAA1671
|
KIAA1671
|
|
chr3_-_150952944
|
0.120
|
NM_016094
|
COMMD2
|
COMM domain containing 2
|
|
chr11_-_104515670
|
0.120
|
NM_021571
|
CARD18
|
caspase recruitment domain family, member 18
|
|
chr11_-_5964790
|
0.119
|
NM_001005173
|
OR52L1
|
olfactory receptor, family 52, subfamily L, member 1
|
|
chrX_+_117410693
|
0.116
|
|
WDR44
|
WD repeat domain 44
|
|
chr11_-_62145952
|
0.113
|
NM_012200
|
B3GAT3
|
beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I)
|
|
chr15_+_55298908
|
0.111
|
NM_207040
|
TCF12
|
transcription factor 12
|
|
chr3_+_99484421
|
0.104
|
NM_001005482
|
OR5H2
|
olfactory receptor, family 5, subfamily H, member 2
|
|
chr1_-_167822248
|
0.104
|
|
F5
|
coagulation factor V (proaccelerin, labile factor)
|
|
chrX_-_77469635
|
0.103
|
NM_006639
|
CYSLTR1
|
cysteinyl leukotriene receptor 1
|
|
chr1_+_245818284
|
0.101
|
NM_001001915
|
OR2G2
|
olfactory receptor, family 2, subfamily G, member 2
|
|
chr9_+_124463665
|
0.100
|
NM_001005236
|
OR1L1
|
olfactory receptor, family 1, subfamily L, member 1
|
|
chr18_+_53967544
|
0.098
|
NM_001144965
NM_001144968
NM_001144969
NM_001144971
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr9_-_21197141
|
0.096
|
NM_002171
|
IFNA10
|
interferon, alpha 10
|
|
chr7_-_107230867
|
0.096
|
NM_000111
|
SLC26A3
|
solute carrier family 26, member 3
|
|
chr7_+_90177082
|
0.095
|
|
CDK14
|
cyclin-dependent kinase 14
|
|
chrX_-_110541029
|
0.095
|
NM_000555
|
DCX
|
doublecortin
|
|
chr12_-_16649211
|
0.094
|
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr7_+_143287952
|
0.093
|
NM_012369
|
OR2F1
|
olfactory receptor, family 2, subfamily F, member 1
|
|
chr3_-_174341653
|
0.092
|
NM_031955
|
SPATA16
|
spermatogenesis associated 16
|
|
chr2_-_43756964
|
0.092
|
NM_001101330
|
LOC728819
|
hCG1645220
|
|
chr8_+_133856738
|
0.091
|
NM_016018
NM_032205
NM_198513
|
PHF20L1
|
PHD finger protein 20-like 1
|
|
chr3_+_82117978
|
0.090
|
|
|
|
|
chr6_+_73133152
|
0.089
|
NM_001168411
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr3_-_150952920
|
0.089
|
|
COMMD2
|
COMM domain containing 2
|
|
chr4_-_120445047
|
0.088
|
NM_001170330
|
C4orf3
|
chromosome 4 open reading frame 3
|
|
chr3_-_56477404
|
0.088
|
NM_015576
|
ERC2
|
ELKS/RAB6-interacting/CAST family member 2
|
|
chr3_-_69120188
|
0.087
|
|
C3orf64
|
chromosome 3 open reading frame 64
|
|
chr1_+_57093030
|
0.086
|
NM_000562
|
C8A
|
complement component 8, alpha polypeptide
|
|
chr1_+_151498802
|
0.085
|
NM_000427
|
LOR
|
loricrin
|
|
chr15_-_74091817
|
0.085
|
NM_138573
|
NRG4
|
neuregulin 4
|
|
chr11_+_71524403
|
0.085
|
NM_000804
|
FOLR3
|
folate receptor 3 (gamma)
|
|
chr7_-_50596238
|
0.085
|
NM_000790
|
DDC
|
dopa decarboxylase (aromatic L-amino acid decarboxylase)
|
|
chr5_+_180726874
|
0.084
|
NM_001005224
NM_001005221
NM_001005277
|
OR4F3
OR4F29
OR4F16
|
olfactory receptor, family 4, subfamily F, member 3
olfactory receptor, family 4, subfamily F, member 29
olfactory receptor, family 4, subfamily F, member 16
|
|
chr3_+_150028130
|
0.084
|
NM_001871
|
CPB1
|
carboxypeptidase B1 (tissue)
|
|
chr15_+_46285789
|
0.084
|
NM_000338
NM_001184832
|
SLC12A1
|
solute carrier family 12 (sodium/potassium/chloride transporters), member 1
|
|
chr11_-_4675524
|
0.084
|
NM_030774
|
OR51E2
|
olfactory receptor, family 51, subfamily E, member 2
|
|
chr18_-_49944986
|
0.082
|
|
MBD2
|
methyl-CpG binding domain protein 2
|
|
chr2_-_210726539
|
0.082
|
|
C2orf67
|
chromosome 2 open reading frame 67
|
|
chr8_+_17874128
|
0.081
|
|
PCM1
|
pericentriolar material 1
|
|
chr3_-_121117647
|
0.081
|
|
GSK3B
|
glycogen synthase kinase 3 beta
|
|
chr1_-_196776697
|
0.081
|
NM_133262
NM_133326
|
ATP6V1G3
|
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G3
|
|
chr11_-_5946299
|
0.080
|
NM_001146033
|
OR56A5
|
olfactory receptor, family 56, subfamily A, member 5
|
|
chr1_+_67404756
|
0.080
|
NM_144701
|
IL23R
|
interleukin 23 receptor
|
|
chr1_-_245237925
|
0.079
|
|
ZNF695
|
zinc finger protein 695
|
|
chr1_-_245237977
|
0.079
|
NM_020394
|
ZNF695
|
zinc finger protein 695
|
|
chr13_-_102209422
|
0.077
|
NM_001146197
|
C13orf40
|
chromosome 13 open reading frame 40
|
|
chr16_-_71639670
|
0.075
|
NM_006885
|
ZFHX3
|
zinc finger homeobox 3
|
|
chrX_+_48221323
|
0.075
|
NM_177434
|
FTSJ1
|
FtsJ homolog 1 (E. coli)
|
|
chr12_+_84792203
|
0.072
|
NM_006183
|
NTS
|
neurotensin
|
|
chr6_+_28032997
|
0.071
|
NM_012367
|
OR2B6
|
olfactory receptor, family 2, subfamily B, member 6
|
|
chr3_+_157321030
|
0.070
|
NM_172160
|
KCNAB1
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1
|
|
chr5_-_58607673
|
0.070
|
NM_001197220
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr4_+_42590038
|
0.069
|
NM_001080476
|
GRXCR1
|
glutaredoxin, cysteine rich 1
|
|
chr4_-_153523113
|
0.068
|
NM_001013415
|
FBXW7
|
F-box and WD repeat domain containing 7
|
|
chr12_-_10453409
|
0.067
|
NM_013431
|
KLRC4-KLRK1
KLRC4
|
KLRC4-KLRK1 read-through transcript
killer cell lectin-like receptor subfamily C, member 4
|
|
chr21_-_36760594
|
0.066
|
NM_012130
|
CLDN14
|
claudin 14
|
|
chr7_+_63411813
|
0.064
|
|
ZNF736
|
zinc finger protein 736
|
|
chr14_+_67238355
|
0.064
|
NM_152443
|
RDH12
|
retinol dehydrogenase 12 (all-trans/9-cis/11-cis)
|
|
chr1_-_176273680
|
0.064
|
|
LOC730102
|
quinone oxidoreductase-like protein 2 pseudogene
|
|
chr6_-_11219932
|
0.064
|
NM_207582
|
ERVFRDE1
|
HERV-FRD provirus ancestral Env polyprotein
|
|
chr4_+_146820805
|
0.064
|
NM_001080531
|
C4orf51
|
chromosome 4 open reading frame 51
|
|
chr6_+_35308605
|
0.064
|
|
SCUBE3
|
signal peptide, CUB domain, EGF-like 3
|
|
chr21_+_30910614
|
0.064
|
NM_181615
|
KRTAP20-1
|
keratin associated protein 20-1
|
|
chr12_+_61645361
|
0.063
|
|
RPL14
|
ribosomal protein L14
|
|
chr1_+_169747788
|
0.063
|
|
PRRC2C
|
proline-rich coiled-coil 2C
|
|
chr17_-_36403910
|
0.062
|
NM_033185
|
KRTAP3-3
|
keratin associated protein 3-3
|
|
chr9_+_4782974
|
0.062
|
|
RCL1
|
RNA terminal phosphate cyclase-like 1
|
|
chr21_-_31175722
|
0.061
|
NM_175858
|
KRTAP11-1
|
keratin associated protein 11-1
|
|
chr12_-_49708279
|
0.060
|
NM_001174125
|
SLC11A2
|
solute carrier family 11 (proton-coupled divalent metal ion transporters), member 2
|
|
chr9_+_92629525
|
0.060
|
NM_001174167
|
SYK
|
spleen tyrosine kinase
|
|
chr10_+_129237602
|
0.059
|
NM_001030013
|
NPS
|
neuropeptide S
|
|
chr12_-_45506001
|
0.059
|
NM_001143824
NM_018018
|
SLC38A4
|
solute carrier family 38, member 4
|
|
chr9_-_21375258
|
0.058
|
NM_000605
|
IFNA2
|
interferon, alpha 2
|
|
chr19_+_44681443
|
0.058
|
|
DLL3
|
delta-like 3 (Drosophila)
|
|
chr8_-_38577891
|
0.058
|
|
|
|
|
chr11_-_3104413
|
0.057
|
|
OSBPL5
|
oxysterol binding protein-like 5
|
|
chr3_+_174598937
|
0.057
|
NM_014932
|
NLGN1
|
neuroligin 1
|
|
chr3_+_112743729
|
0.056
|
|
CD96
|
CD96 molecule
|
|
chr20_+_19818209
|
0.056
|
NM_018993
|
RIN2
|
Ras and Rab interactor 2
|
|
chr7_+_44612645
|
0.056
|
NM_001003941
NM_001165036
NM_002541
|
OGDH
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide)
|
|
chr1_+_13903936
|
0.056
|
NM_012231
NM_015866
|
PRDM2
|
PR domain containing 2, with ZNF domain
|
|
chr20_+_33223434
|
0.055
|
NM_006404
|
PROCR
|
protein C receptor, endothelial
|
|
chr12_+_76749199
|
0.054
|
NM_014903
|
NAV3
|
neuron navigator 3
|
|
chr3_+_153014550
|
0.054
|
NM_001086
|
AADAC
|
arylacetamide deacetylase (esterase)
|
|
chr5_+_140583098
|
0.053
|
NM_018934
|
PCDHB14
|
protocadherin beta 14
|
|
chr11_-_47502115
|
0.053
|
NM_001172639
|
CELF1
|
CUGBP, Elav-like family member 1
|
|
chr1_-_153477511
|
0.052
|
|
GBA
|
glucosidase, beta, acid
|
|
chr5_-_179040580
|
0.052
|
NM_001164444
|
CBY3
|
chibby homolog 3 (Drosophila)
|
|
chr12_-_119249938
|
0.051
|
NM_000928
|
PLA2G1B
|
phospholipase A2, group IB (pancreas)
|
|
chr8_+_133856793
|
0.051
|
|
PHF20L1
|
PHD finger protein 20-like 1
|
|
chr5_+_140730014
|
0.051
|
NM_018924
NM_032097
|
PCDHGB3
|
protocadherin gamma subfamily B, 3
|
|
chr22_+_29696662
|
0.051
|
|
|
|
|
chr9_-_14076901
|
0.051
|
|
NFIB
|
nuclear factor I/B
|
|
chr4_+_68995761
|
0.050
|
NM_014058
|
TMPRSS11E
|
transmembrane protease, serine 11E
|
|
chr1_-_156056456
|
0.050
|
NM_001159397
NM_001159398
NM_052938
|
FCRL1
|
Fc receptor-like 1
|
|
chr10_+_89409332
|
0.050
|
NM_001015880
NM_004670
|
PAPSS2
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2
|
|
chr3_+_112743615
|
0.050
|
NM_005816
NM_198196
|
CD96
|
CD96 molecule
|
|
chr7_+_91762227
|
0.049
|
|
|
|
|
chr4_-_40631397
|
0.048
|
|
APBB2
|
amyloid beta (A4) precursor protein-binding, family B, member 2
|
|
chr12_-_119826433
|
0.048
|
NM_139015
|
SPPL3
|
signal peptide peptidase 3
|
|
chr19_-_43926953
|
0.048
|
NM_144691
|
CAPN12
|
calpain 12
|
|
chr12_-_85756766
|
0.047
|
NM_013244
|
MGAT4C
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative)
|
|
chr20_-_44575599
|
0.047
|
NM_018102
NM_199441
|
ZNF334
|
zinc finger protein 334
|
|
chr6_+_25760352
|
0.047
|
NM_006998
|
SCGN
|
secretagogin, EF-hand calcium binding protein
|
|
chr3_-_126576693
|
0.046
|
NM_021964
|
ZNF148
|
zinc finger protein 148
|
|
chr14_+_19598043
|
0.045
|
NM_001004717
|
OR4L1
|
olfactory receptor, family 4, subfamily L, member 1
|
|
chrX_+_27736027
|
0.045
|
NM_182506
|
MAGEB10
|
melanoma antigen family B, 10
|
|
chr3_-_194579207
|
0.045
|
NM_198505
|
ATP13A5
|
ATPase type 13A5
|
|
chr12_-_44670584
|
0.044
|
|
SCAF11
|
SR-related CTD-associated factor 11
|
|
chr1_-_84099261
|
0.044
|
|
|
|
|
chr3_+_123279439
|
0.044
|
NM_006889
|
CD86
|
CD86 molecule
|
|
chr3_-_87408301
|
0.044
|
NM_000306
NM_001122757
|
POU1F1
|
POU class 1 homeobox 1
|
|
chr8_-_42817604
|
0.043
|
NM_018105
NM_199003
|
THAP1
|
THAP domain containing, apoptosis associated protein 1
|
|
chr12_-_66905802
|
0.043
|
NM_018402
|
IL26
|
interleukin 26
|