|
chr8_+_32698892
|
2.912
|
NM_001159996
|
NRG1
|
neuregulin 1
|
|
chr4_-_153523113
|
2.630
|
NM_001013415
|
FBXW7
|
F-box and WD repeat domain containing 7
|
|
chr11_+_101488380
|
2.619
|
NM_001195045
|
YAP1
|
Yes-associated protein 1
|
|
chr3_+_99465777
|
2.590
|
NM_001005479
|
OR5H6
|
olfactory receptor, family 5, subfamily H, member 6
|
|
chr6_+_127940011
|
2.455
|
NM_001010905
|
C6orf58
|
chromosome 6 open reading frame 58
|
|
chr14_-_105862080
|
2.444
|
|
IGHV4-31
IGHA1
IGHD
IGHG1
IGHG2
IGHG3
IGHM
SCFV
|
immunoglobulin heavy variable 4-31
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant delta
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant gamma 2 (G2m marker)
immunoglobulin heavy constant gamma 3 (G3m marker)
immunoglobulin heavy constant mu
single-chain Fv fragment
|
|
chr14_-_106064887
|
2.266
|
|
LOC100126583
IGHV3-48
IGHA1
IGHM
|
hypothetical LOC100126583
immunoglobulin heavy variable 3-48
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant mu
|
|
chr14_+_38719462
|
2.240
|
|
PNN
|
pinin, desmosome associated protein
|
|
chr7_+_93388951
|
2.183
|
NM_004126
|
GNG11
|
guanine nucleotide binding protein (G protein), gamma 11
|
|
chr1_+_150852910
|
2.100
|
NM_178433
|
LCE3B
|
late cornified envelope 3B
|
|
chr2_+_228386800
|
2.005
|
NM_001130046
NM_004591
|
CCL20
|
chemokine (C-C motif) ligand 20
|
|
chr6_-_109795925
|
1.967
|
|
CD164
|
CD164 molecule, sialomucin
|
|
chr14_+_38720085
|
1.957
|
|
PNN
|
pinin, desmosome associated protein
|
|
chr6_+_153113624
|
1.953
|
NM_003381
NM_194435
|
VIP
|
vasoactive intestinal peptide
|
|
chr7_+_17304707
|
1.902
|
NM_001621
|
AHR
|
aryl hydrocarbon receptor
|
|
chr14_+_31615959
|
1.846
|
|
ARHGAP5
|
Rho GTPase activating protein 5
|
|
chr2_-_215967442
|
1.838
|
|
FN1
|
fibronectin 1
|
|
chr9_-_106401514
|
1.832
|
NM_001004482
|
OR13C5
|
olfactory receptor, family 13, subfamily C, member 5
|
|
chr14_+_64001969
|
1.826
|
NM_004857
|
AKAP5
|
A kinase (PRKA) anchor protein 5
|
|
chr4_+_95595419
|
1.756
|
|
PDLIM5
|
PDZ and LIM domain 5
|
|
chr11_-_95715976
|
1.679
|
NM_032427
|
MAML2
|
mastermind-like 2 (Drosophila)
|
|
chr4_+_124540114
|
1.673
|
NM_005841
|
SPRY1
|
sprouty homolog 1, antagonist of FGF signaling (Drosophila)
|
|
chr2_-_210888010
|
1.665
|
NM_079420
|
MYL1
|
myosin, light chain 1, alkali; skeletal, fast
|
|
chr14_+_31615922
|
1.657
|
|
ARHGAP5
|
Rho GTPase activating protein 5
|
|
chr13_+_81162030
|
1.640
|
|
|
|
|
chr5_-_146441184
|
1.639
|
NM_181677
NM_181678
|
PPP2R2B
|
protein phosphatase 2, regulatory subunit B, beta
|
|
chr14_-_20969706
|
1.605
|
NM_001170629
|
CHD8
|
chromodomain helicase DNA binding protein 8
|
|
chr12_-_11178109
|
1.593
|
NM_001097643
|
TAS2R30
|
taste receptor, type 2, member 30
|
|
chr6_+_88024152
|
1.591
|
|
ZNF292
|
zinc finger protein 292
|
|
chr14_-_94669547
|
1.569
|
NM_001195573
|
DICER1
|
dicer 1, ribonuclease type III
|
|
chr18_+_27281729
|
1.549
|
NM_001944
|
DSG3
|
desmoglein 3
|
|
chr5_+_27508139
|
1.533
|
|
LOC643401
|
hypothetical protein LOC643401
|
|
chr6_-_127882192
|
1.532
|
NM_001012279
|
C6orf174
|
chromosome 6 open reading frame 174
|
|
chr2_+_191009265
|
1.532
|
|
MFSD6
|
major facilitator superfamily domain containing 6
|
|
chr14_-_105589467
|
1.528
|
|
LOC100126583
IGHA1
IGHG1
IGHG2
IGHM
|
hypothetical LOC100126583
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant gamma 2 (G2m marker)
immunoglobulin heavy constant mu
|
|
chr2_-_225073357
|
1.519
|
|
CUL3
|
cullin 3
|
|
chr6_+_129245978
|
1.515
|
NM_000426
NM_001079823
|
LAMA2
|
laminin, alpha 2
|
|
chr9_+_12764988
|
1.508
|
NM_203403
|
C9orf150
|
chromosome 9 open reading frame 150
|
|
chr9_-_21375258
|
1.495
|
NM_000605
|
IFNA2
|
interferon, alpha 2
|
|
chr10_+_112669353
|
1.495
|
|
SHOC2
|
soc-2 suppressor of clear homolog (C. elegans)
|
|
chr10_+_90336489
|
1.489
|
NM_001010939
|
LIPJ
|
lipase, family member J
|
|
chr14_-_106193403
|
1.484
|
|
IGKV3-20
|
immunoglobulin kappa variable 3-20
|
|
chr14_+_22059212
|
1.474
|
|
|
|
|
chr7_+_80145461
|
1.467
|
|
CD36
|
CD36 molecule (thrombospondin receptor)
|
|
chr11_+_27019084
|
1.450
|
NM_003986
|
BBOX1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1
|
|
chr3_+_186563529
|
1.447
|
NM_004721
|
MAP3K13
|
mitogen-activated protein kinase kinase kinase 13
|
|
chr2_-_40593074
|
1.438
|
NM_001112802
|
SLC8A1
|
solute carrier family 8 (sodium/calcium exchanger), member 1
|
|
chr2_+_114390975
|
1.425
|
|
ACTR3
|
ARP3 actin-related protein 3 homolog (yeast)
|
|
chr10_+_112669349
|
1.384
|
|
SHOC2
|
soc-2 suppressor of clear homolog (C. elegans)
|
|
chr17_-_36451189
|
1.380
|
NM_030967
|
KRTAP1-3
KRTAP1-1
|
keratin associated protein 1-3
keratin associated protein 1-1
|
|
chr1_-_246503738
|
1.379
|
NM_001004695
|
OR2T33
|
olfactory receptor, family 2, subfamily T, member 33
|
|
chr1_-_246554492
|
1.360
|
NM_001004691
|
OR2M7
|
olfactory receptor, family 2, subfamily M, member 7
|
|
chr17_-_26648375
|
1.360
|
NM_002544
|
OMG
|
oligodendrocyte myelin glycoprotein
|
|
chr7_-_22518200
|
1.352
|
|
EEF1A1
|
eukaryotic translation elongation factor 1 alpha 1
|
|
chr12_-_51153788
|
1.351
|
NM_173086
|
KRT6C
|
keratin 6C
|
|
chr6_-_68655708
|
1.349
|
|
|
|
|
chr9_+_112470871
|
1.341
|
NM_001166280
NM_001166281
NM_005592
|
MUSK
|
muscle, skeletal, receptor tyrosine kinase
|
|
chr5_-_147191428
|
1.340
|
NM_003122
|
SPINK1
|
serine peptidase inhibitor, Kazal type 1
|
|
chr1_-_150564302
|
1.337
|
NM_002016
|
FLG
|
filaggrin
|
|
chr7_+_94135384
|
1.333
|
|
PEG10
|
paternally expressed 10
|
|
chr20_-_17589122
|
1.324
|
|
RRBP1
|
ribosome binding protein 1 homolog 180kDa (dog)
|
|
chr17_-_36376663
|
1.320
|
NM_213656
|
KRT39
|
keratin 39
|
|
chr18_+_30544193
|
1.317
|
NM_001198941
NM_032978
|
DTNA
|
dystrobrevin, alpha
|
|
chr9_-_72673753
|
1.312
|
NM_001007470
NM_020952
NM_024971
NM_206944
NM_206945
NM_206946
NM_206947
NM_206948
|
TRPM3
|
transient receptor potential cation channel, subfamily M, member 3
|
|
chr14_-_31026901
|
1.299
|
NM_001197184
|
GPR33
|
G protein-coupled receptor 33 (gene/pseudogene)
|
|
chr11_-_89248832
|
1.297
|
NM_001164397
|
TRIM64B
|
tripartite motif containing 64B
|
|
chr9_-_21358005
|
1.263
|
NM_006900
|
IFNA1
IFNA13
|
interferon, alpha 1
interferon, alpha 13
|
|
chr14_+_74353417
|
1.259
|
|
YLPM1
|
YLP motif containing 1
|
|
chr5_-_86739683
|
1.257
|
|
CCNH
|
cyclin H
|
|
chr5_-_147191334
|
1.253
|
|
SPINK1
|
serine peptidase inhibitor, Kazal type 1
|
|
chr10_+_63092724
|
1.251
|
NM_173554
|
C10orf107
|
chromosome 10 open reading frame 107
|
|
chr14_+_55148488
|
1.249
|
|
KTN1
|
kinectin 1 (kinesin receptor)
|
|
chr6_+_53043729
|
1.249
|
NM_012347
|
FBXO9
|
F-box protein 9
|
|
chr11_-_5178505
|
1.238
|
NM_001004760
|
OR51V1
|
olfactory receptor, family 51, subfamily V, member 1
|
|
chr13_+_49658016
|
1.234
|
|
|
|
|
chr10_-_98280299
|
1.210
|
|
TM9SF3
|
transmembrane 9 superfamily member 3
|
|
chr13_-_32678142
|
1.208
|
NM_052851
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr6_+_64414389
|
1.201
|
NM_015153
|
PHF3
|
PHD finger protein 3
|
|
chr12_-_11075272
|
1.195
|
NM_176885
|
TAS2R31
|
taste receptor, type 2, member 31
|
|
chr1_+_39569396
|
1.195
|
NM_033044
|
MACF1
|
microtubule-actin crosslinking factor 1
|
|
chr14_-_106184988
|
1.194
|
|
IGHA1
IGHG1
|
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr11_+_56187713
|
1.190
|
NM_001004730
|
OR5AR1
|
olfactory receptor, family 5, subfamily AR, member 1
|
|
chr1_+_93317374
|
1.187
|
NM_001164391
NM_001164392
NM_001164393
NM_007358
|
MTF2
|
metal response element binding transcription factor 2
|
|
chr2_+_192251105
|
1.186
|
NM_001031716
|
OBFC2A
|
oligonucleotide/oligosaccharide-binding fold containing 2A
|
|
chr7_+_111879348
|
1.184
|
NM_001197079
|
IFRD1
|
interferon-related developmental regulator 1
|
|
chr11_+_89341319
|
1.184
|
NM_001136486
|
TRIM64
|
tripartite motif containing 64
|
|
chr9_+_122890394
|
1.178
|
NM_007018
|
CEP110
|
centrosomal protein 110kDa
|
|
chr1_+_157067729
|
1.174
|
NM_002432
|
MNDA
|
myeloid cell nuclear differentiation antigen
|
|
chr10_-_113933457
|
1.172
|
NM_020918
|
GPAM
|
glycerol-3-phosphate acyltransferase, mitochondrial
|
|
chr10_-_121592020
|
1.171
|
|
MCMBP
|
minichromosome maintenance complex binding protein
|
|
chr14_+_22059174
|
1.169
|
|
|
|
|
chr14_-_105763152
|
1.168
|
|
IGHV4-31
IGHA1
IGHD
IGHG1
IGHG3
|
immunoglobulin heavy variable 4-31
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant delta
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant gamma 3 (G3m marker)
|
|
chr10_+_86121526
|
1.161
|
|
FAM190B
|
family with sequence similarity 190, member B
|
|
chr5_+_65476418
|
1.161
|
NM_139168
|
SREK1
|
splicing regulatory glutamine/lysine-rich protein 1
|
|
chr22_+_29696662
|
1.160
|
|
|
|
|
chr4_-_122357104
|
1.159
|
NM_001128843
|
TNIP3
|
TNFAIP3 interacting protein 3
|
|
chr12_-_11041740
|
1.155
|
NM_176889
|
TAS2R20
|
taste receptor, type 2, member 20
|
|
chr4_-_120462705
|
1.155
|
NM_000134
|
FABP2
|
fatty acid binding protein 2, intestinal
|
|
chr5_+_140160966
|
1.155
|
NM_018906
NM_031497
|
PCDHA3
|
protocadherin alpha 3
|
|
chr18_+_3443771
|
1.150
|
NM_173211
|
TGIF1
|
TGFB-induced factor homeobox 1
|
|
chr19_+_41419165
|
1.141
|
|
ZNF146
|
zinc finger protein 146
|
|
chr3_+_180348818
|
1.140
|
NM_006218
|
PIK3CA
|
phosphoinositide-3-kinase, catalytic, alpha polypeptide
|
|
chr21_+_30937053
|
1.136
|
NM_001128077
|
KRTAP20-3
|
keratin associated protein 20-3
|
|
chr5_-_101862447
|
1.124
|
NM_173488
|
SLCO6A1
|
solute carrier organic anion transporter family, member 6A1
|
|
chr13_+_19430881
|
1.120
|
|
ZMYM2
|
zinc finger, MYM-type 2
|
|
chr13_-_101852028
|
1.113
|
NM_175929
|
FGF14
|
fibroblast growth factor 14
|
|
chr14_+_98247702
|
1.108
|
NM_182560
|
C14orf177
|
chromosome 14 open reading frame 177
|
|
chr5_+_43645018
|
1.104
|
|
NNT
|
nicotinamide nucleotide transhydrogenase
|
|
chr11_+_33306652
|
1.103
|
|
HIPK3
|
homeodomain interacting protein kinase 3
|
|
chr1_-_115070413
|
1.103
|
|
CSDE1
|
cold shock domain containing E1, RNA-binding
|
|
chr2_-_231697999
|
1.094
|
NM_000867
|
HTR2B
|
5-hydroxytryptamine (serotonin) receptor 2B
|
|
chr12_-_12182641
|
1.086
|
|
LRP6
|
low density lipoprotein receptor-related protein 6
|
|
chrX_+_56772391
|
1.086
|
|
LOC550643
|
hypothetical LOC550643
|
|
chr11_+_11820096
|
1.085
|
|
USP47
|
ubiquitin specific peptidase 47
|
|
chr21_-_26294332
|
1.084
|
|
APP
|
amyloid beta (A4) precursor protein
|
|
chr1_-_196776697
|
1.078
|
NM_133262
NM_133326
|
ATP6V1G3
|
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G3
|
|
chr11_-_6005546
|
1.078
|
NM_001001917
|
OR56A1
|
olfactory receptor, family 56, subfamily A, member 1
|
|
chr4_-_101658094
|
1.070
|
NM_001159694
NM_016242
|
EMCN
|
endomucin
|
|
chr5_-_151764857
|
1.068
|
NM_020167
|
NMUR2
|
neuromedin U receptor 2
|
|
chr6_-_11887265
|
1.068
|
NM_001143948
NM_032744
|
C6orf105
|
chromosome 6 open reading frame 105
|
|
chr9_+_124416768
|
1.063
|
NM_012364
|
OR1Q1
|
olfactory receptor, family 1, subfamily Q, member 1
|
|
chr4_+_144654526
|
1.062
|
|
|
|
|
chr10_+_112669267
|
1.061
|
NM_007373
|
SHOC2
|
soc-2 suppressor of clear homolog (C. elegans)
|
|
chr17_+_36642240
|
1.060
|
NM_031962
|
KRTAP9-9
KRTAP9-3
|
keratin associated protein 9-9
keratin associated protein 9-3
|
|
chr13_+_48448950
|
1.060
|
|
FNDC3A
|
fibronectin type III domain containing 3A
|
|
chr14_-_20694023
|
1.058
|
NM_001004731
|
OR5AU1
|
olfactory receptor, family 5, subfamily AU, member 1
|
|
chr22_-_39189369
|
1.047
|
|
MKL1
|
megakaryoblastic leukemia (translocation) 1
|
|
chr18_-_35634247
|
1.045
|
|
LOC647946
|
hypothetical LOC647946
|
|
chr13_+_48448912
|
1.042
|
|
FNDC3A
|
fibronectin type III domain containing 3A
|
|
chr12_+_58369392
|
1.042
|
NM_004731
|
SLC16A7
|
solute carrier family 16, member 7 (monocarboxylic acid transporter 2)
|
|
chr1_-_246823691
|
1.040
|
NM_001004693
|
OR2T10
|
olfactory receptor, family 2, subfamily T, member 10
|
|
chr9_+_74956494
|
1.035
|
NM_000700
|
ANXA1
|
annexin A1
|
|
chr13_+_48582389
|
1.034
|
NM_014923
|
FNDC3A
|
fibronectin type III domain containing 3A
|
|
chr7_-_14847479
|
1.032
|
NM_004080
NM_145695
|
DGKB
|
diacylglycerol kinase, beta 90kDa
|
|
chr3_-_52688768
|
1.030
|
NM_018165
NM_181042
|
PBRM1
|
polybromo 1
|
|
chr12_+_77963563
|
1.022
|
NM_001135806
|
SYT1
|
synaptotagmin I
|
|
chr2_-_21079145
|
1.017
|
|
APOB
|
apolipoprotein B (including Ag(x) antigen)
|
|
chr6_+_132901119
|
1.016
|
NM_175057
|
TAAR9
|
trace amine associated receptor 9 (gene/pseudogene)
|
|
chr10_+_129237602
|
1.016
|
NM_001030013
|
NPS
|
neuropeptide S
|
|
chr3_+_73193499
|
1.012
|
NM_018029
|
EBLN2
|
endogenous Borna-like N element-2
|
|
chr21_-_30720100
|
1.007
|
NM_181622
|
KRTAP13-3
|
keratin associated protein 13-3
|
|
chr8_-_82522175
|
1.007
|
NM_002677
|
PMP2
|
peripheral myelin protein 2
|
|
chr10_+_106003941
|
1.006
|
NM_001191003
|
GSTO1
|
glutathione S-transferase omega 1
|
|
chr17_+_23394062
|
1.005
|
|
NLK
|
nemo-like kinase
|
|
chr7_+_93862137
|
1.005
|
|
COL1A2
PLAG1
|
collagen, type I, alpha 2
pleiomorphic adenoma gene 1
|
|
chr13_+_48448628
|
1.001
|
|
FNDC3A
|
fibronectin type III domain containing 3A
|
|
chr6_-_128883125
|
1.001
|
NM_002844
NM_001135648
|
PTPRK
|
protein tyrosine phosphatase, receptor type, K
|
|
chr6_-_26343139
|
0.994
|
NM_005320
|
HIST1H1D
|
histone cluster 1, H1d
|
|
chr17_-_3142625
|
0.993
|
NM_002550
|
OR3A1
|
olfactory receptor, family 3, subfamily A, member 1
|
|
chr21_-_26345294
|
0.990
|
|
|
|
|
chr9_-_114814286
|
0.988
|
NM_001101338
|
ZNF883
|
zinc finger protein 883
|
|
chr3_-_71715431
|
0.986
|
NM_001012505
NM_032682
|
FOXP1
|
forkhead box P1
|
|
chr2_-_213723298
|
0.986
|
NM_016260
|
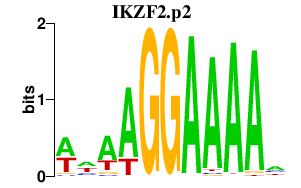
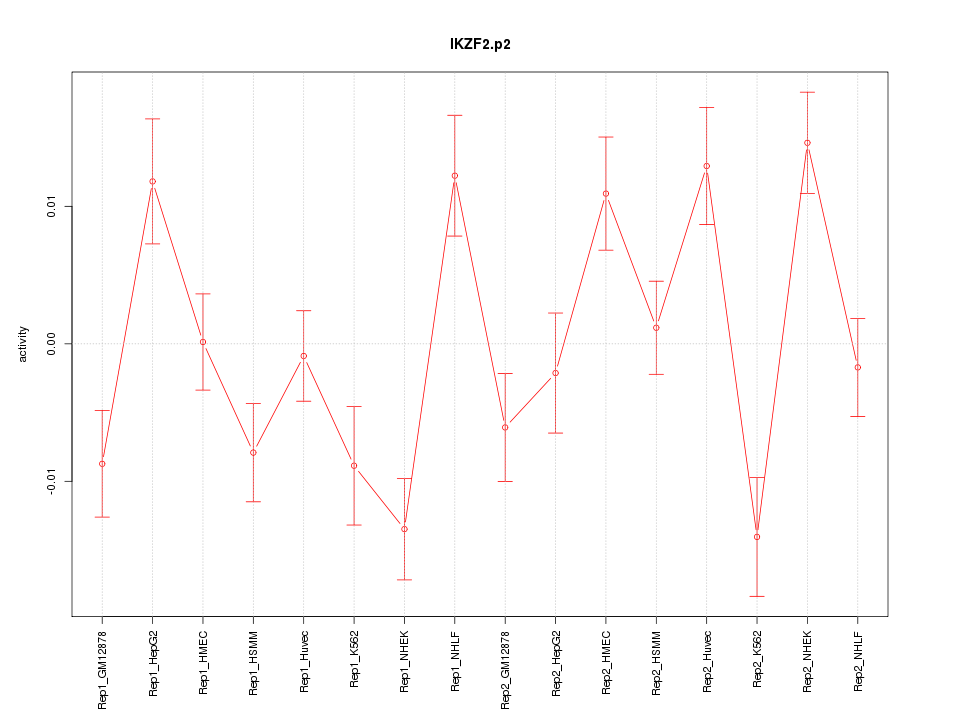
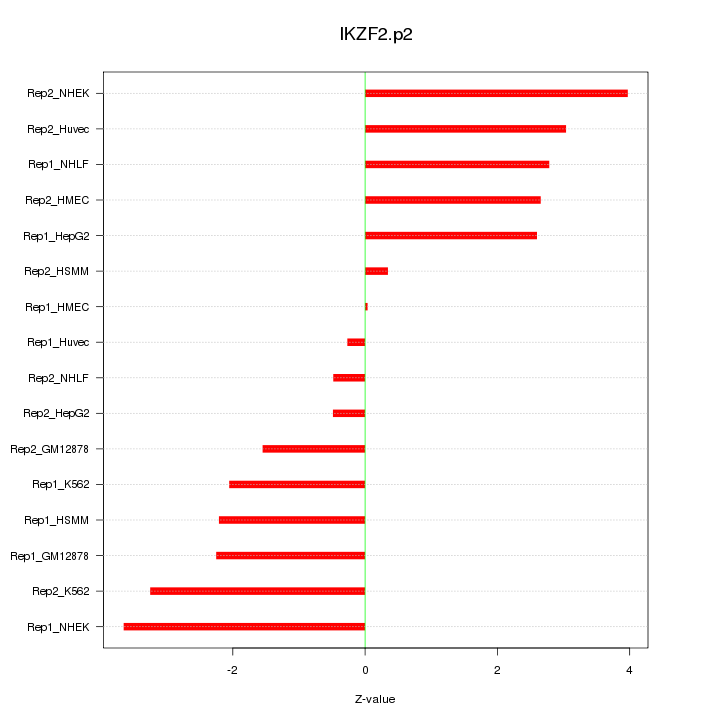
IKZF2
|
IKAROS family zinc finger 2 (Helios)
|
|
chr9_-_28709302
|
0.985
|
NM_152570
|
LINGO2
|
leucine rich repeat and Ig domain containing 2
|
|
chr12_-_88573949
|
0.978
|
NM_001001323
NM_001682
|
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1
|
|
chr10_+_96433240
|
0.973
|
NM_000772
NM_001128925
|
CYP2C18
|
cytochrome P450, family 2, subfamily C, polypeptide 18
|
|
chr10_-_98311813
|
0.972
|
|
TM9SF3
|
transmembrane 9 superfamily member 3
|
|
chr14_-_105785639
|
0.971
|
|
|
|
|
chr1_-_21310462
|
0.962
|
NM_003760
|
EIF4G3
|
eukaryotic translation initiation factor 4 gamma, 3
|
|
chr16_+_31105579
|
0.959
|
|
|
|
|
chr12_+_7348194
|
0.955
|
NM_001080454
|
ACSM4
|
acyl-CoA synthetase medium-chain family member 4
|
|
chr8_+_120289735
|
0.952
|
NM_052886
|
MAL2
|
mal, T-cell differentiation protein 2 (gene/pseudogene)
|
|
chr4_+_25931745
|
0.944
|
|
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region
|
|
chr13_+_23042722
|
0.943
|
NM_148957
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19
|
|
chr11_+_55776251
|
0.941
|
NM_001004747
|
OR5T3
|
olfactory receptor, family 5, subfamily T, member 3
|
|
chr1_-_246869181
|
0.926
|
NM_001001827
|
OR2T35
|
olfactory receptor, family 2, subfamily T, member 35
|
|
chr10_+_102285690
|
0.924
|
|
HIF1AN
|
hypoxia inducible factor 1, alpha subunit inhibitor
|
|
chr2_-_189363075
|
0.924
|
NM_052952
|
DIRC1
|
disrupted in renal carcinoma 1
|
|
chr1_+_42934910
|
0.923
|
|
YBX1
|
Y box binding protein 1
|
|
chr17_+_12564330
|
0.921
|
NM_001146313
|
MYOCD
|
myocardin
|
|
chr11_+_113815317
|
0.920
|
NM_015523
|
REXO2
|
REX2, RNA exonuclease 2 homolog (S. cerevisiae)
|
|
chr9_-_14900992
|
0.920
|
NM_144966
|
FREM1
|
FRAS1 related extracellular matrix 1
|
|
chr17_+_36636404
|
0.920
|
NM_031961
|
KRTAP9-2
|
keratin associated protein 9-2
|
|
chr7_+_91762227
|
0.916
|
|
|
|
|
chr5_+_140235114
|
0.914
|
NM_018903
NM_031864
|
PCDHA12
|
protocadherin alpha 12
|
|
chr18_-_46600295
|
0.914
|
NM_001127175
NM_001127176
NM_031939
|
MRO
|
maestro
|
|
chr10_-_112668679
|
0.913
|
NM_001195306
|
BBIP1
|
BBSome interacting protein 1
|
|
chr9_+_70046658
|
0.913
|
NM_201453
|
CBWD5
CBWD3
CBWD1
CBWD6
|
COBW domain containing 5
COBW domain containing 3
COBW domain containing 1
COBW domain containing 6
|
|
chr9_+_108665168
|
0.912
|
NM_021224
|
ZNF462
|
zinc finger protein 462
|
|
chr2_-_49235059
|
0.906
|
NM_000145
NM_181446
|
FSHR
|
follicle stimulating hormone receptor
|
|
chr9_-_21192201
|
0.905
|
NM_021057
|
IFNA7
|
interferon, alpha 7
|
|
chr7_-_18033970
|
0.905
|
NM_175886
|
PRPS1L1
|
phosphoribosyl pyrophosphate synthetase 1-like 1
|
|
chr17_+_23393744
|
0.903
|
NM_016231
|
NLK
|
nemo-like kinase
|
|
chr14_+_85066265
|
0.903
|
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2
|
|
chr16_+_10430225
|
0.893
|
NM_024997
|
ATF7IP2
|
activating transcription factor 7 interacting protein 2
|
|
chr11_+_55297489
|
0.888
|
NM_001001967
|
OR5D13
|
olfactory receptor, family 5, subfamily D, member 13
|
|
chr7_-_122627235
|
0.886
|
NM_022444
|
SLC13A1
|
solute carrier family 13 (sodium/sulfate symporters), member 1
|
|
chr15_-_75259714
|
0.882
|
|
PEAK1
|
NKF3 kinase family member
|
|
chr14_-_22549168
|
0.882
|
NM_001130706
NM_001130708
NM_021944
|
C14orf93
|
chromosome 14 open reading frame 93
|
|
chr1_-_171838802
|
0.882
|
NM_178527
|
SLC9A11
|
solute carrier family 9, member 11
|
|
chr4_+_71054492
|
0.873
|
NM_214711
|
C4orf40
|
chromosome 4 open reading frame 40
|
|
chr1_+_40399618
|
0.871
|
NM_012421
|
RLF
|
rearranged L-myc fusion
|
|
chr8_-_17812151
|
0.869
|
|
|
|
|
chr20_-_33783409
|
0.868
|
|
RBM39
|
RNA binding motif protein 39
|
|
chr11_+_109468734
|
0.865
|
|
ZC3H12C
|
zinc finger CCCH-type containing 12C
|
|
chr1_+_40399642
|
0.862
|
|
RLF
|
rearranged L-myc fusion
|
|
chr4_+_74520796
|
0.862
|
NM_001134
|
AFP
|
alpha-fetoprotein
|
|
chr2_-_210876560
|
0.861
|
NM_079422
|
MYL1
|
myosin, light chain 1, alkali; skeletal, fast
|
|
chr19_+_41418723
|
0.856
|
|
ZNF146
|
zinc finger protein 146
|
|
chr11_-_62753574
|
0.854
|
NM_199352
|
SLC22A25
|
solute carrier family 22, member 25
|