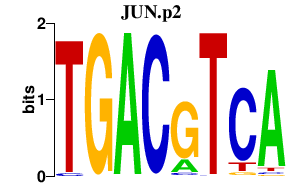
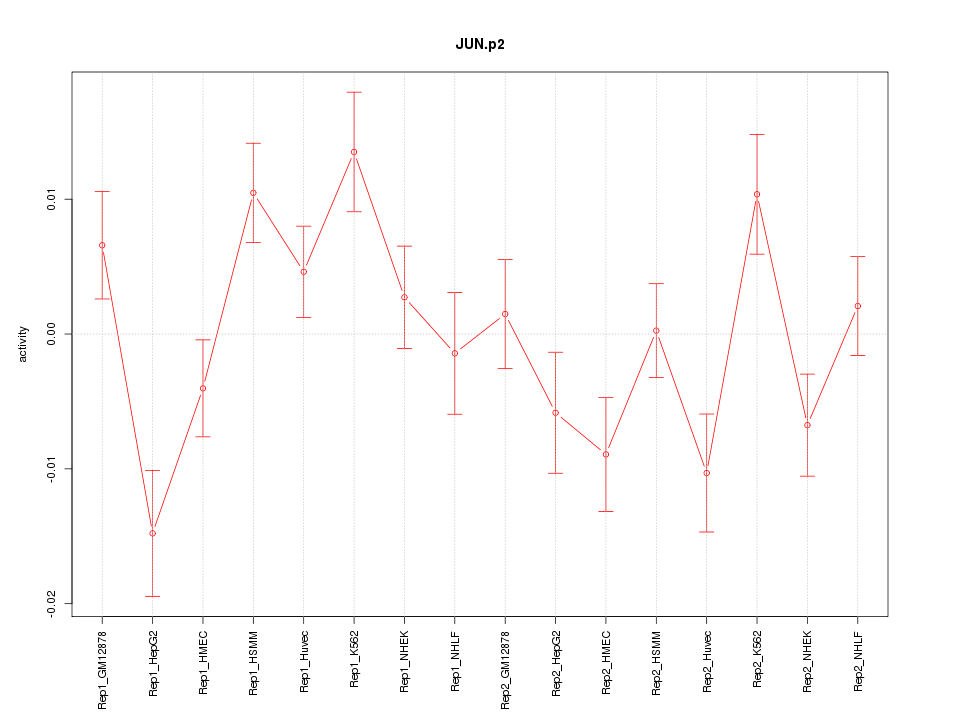
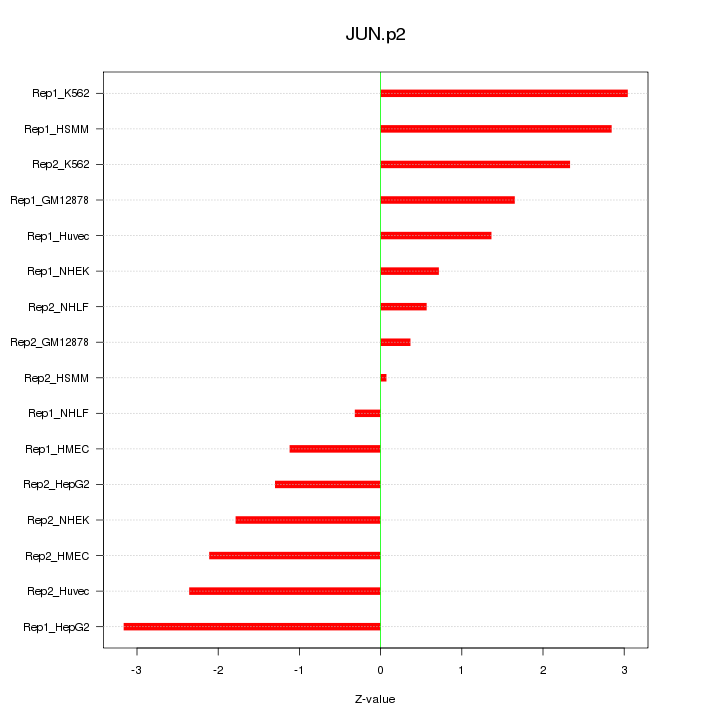
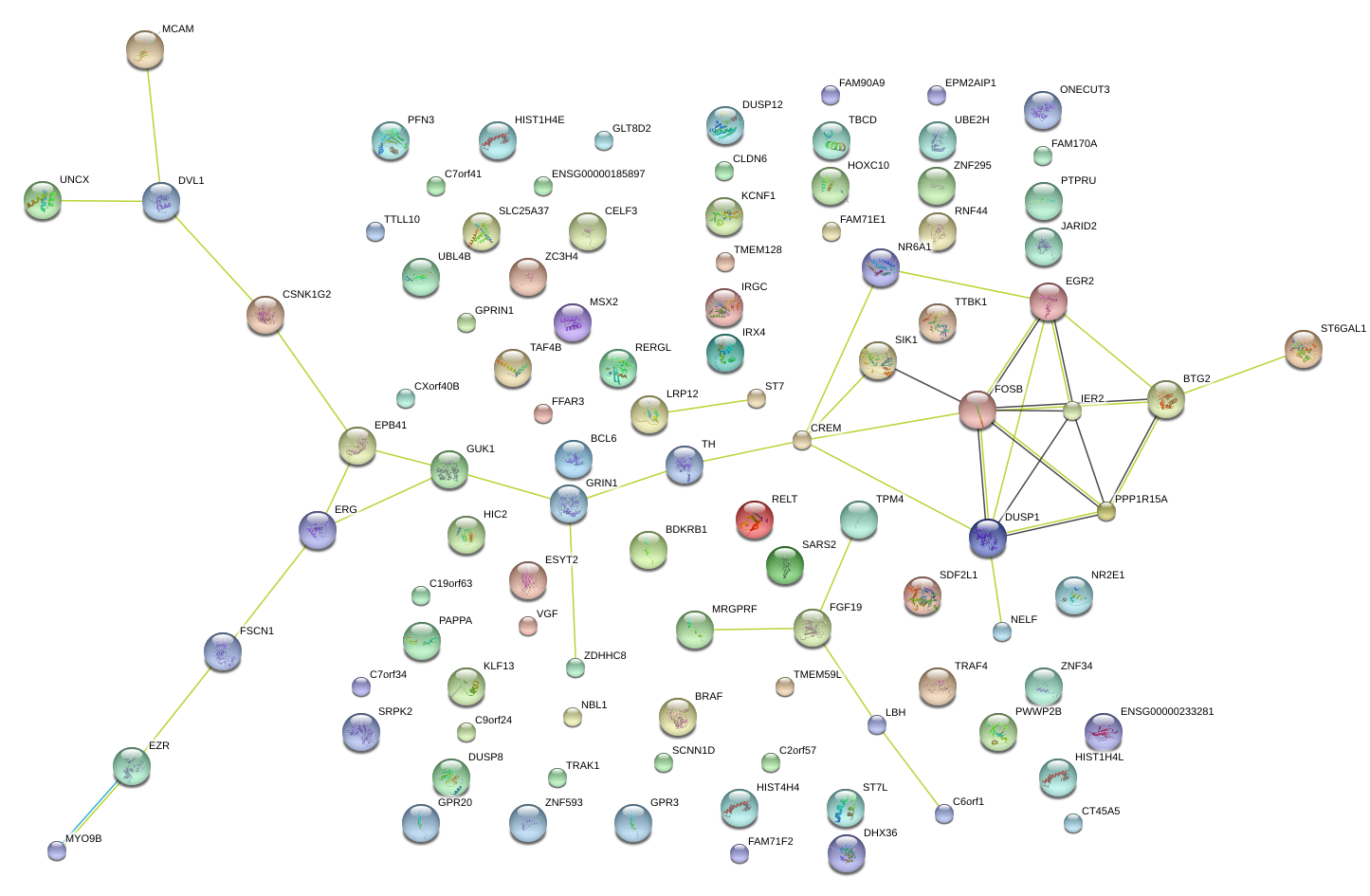
|
chr1_+_26368969
|
5.502
|
NM_015871
|
ZNF593
|
zinc finger protein 593
|
|
chr21_-_43671355
|
4.916
|
NM_173354
|
SIK1
|
salt-inducible kinase 1
|
|
chr22_+_20101661
|
4.399
|
NM_015094
|
HIC2
|
hypermethylated in cancer 2
|
|
chr19_+_50663089
|
3.473
|
NM_001114171
NM_006732
|
FOSB
|
FBJ murine osteosarcoma viral oncogene homolog B
|
|
chr12_+_52665196
|
3.277
|
NM_017409
|
HOXC10
|
homeobox C10
|
|
chr6_+_43319199
|
3.194
|
NM_032538
|
TTBK1
|
tau tubulin kinase 1
|
|
chr7_-_100595552
|
2.838
|
NM_003378
|
VGF
|
VGF nerve growth factor inducible
|
|
chr8_+_23442331
|
2.654
|
|
SLC25A37
|
solute carrier family 25, member 37
|
|
chr1_+_29435534
|
2.515
|
NM_001195001
NM_005704
NM_133177
NM_133178
|
PTPRU
|
protein tyrosine phosphatase, receptor type, U
|
|
chr8_-_142446546
|
2.464
|
NM_005293
|
GPR20
|
G protein-coupled receptor 20
|
|
chr7_-_129379947
|
2.434
|
NM_003344
NM_182697
|
UBE2H
|
ubiquitin-conjugating enzyme E2H (UBC8 homolog, yeast)
|
|
chr10_+_134060681
|
2.413
|
NM_001098637
NM_138499
|
PWWP2B
|
PWWP domain containing 2B
|
|
chr8_+_23442413
|
2.262
|
|
SLC25A37
|
solute carrier family 25, member 37
|
|
chr11_-_2149591
|
2.246
|
NM_000360
NM_199292
NM_199293
|
TH
|
tyrosine hydroxylase
|
|
chr3_+_42165740
|
2.241
|
|
TRAK1
|
trafficking protein, kinesin binding 1
|
|
chr7_+_116447499
|
2.116
|
|
ST7
|
suppression of tumorigenicity 7
|
|
chr19_+_16048266
|
2.029
|
|
TPM4
|
tropomyosin 4
|
|
chr19_+_16048315
|
2.021
|
|
TPM4
|
tropomyosin 4
|
|
chr21_-_42303517
|
1.955
|
NM_001098402
NM_001098403
NM_020727
|
ZNF295
|
zinc finger protein 295
|
|
chr7_+_142346724
|
1.894
|
NM_178829
|
C7orf34
|
chromosome 7 open reading frame 34
|
|
chr9_+_139152595
|
1.892
|
|
GRIN1
|
glutamate receptor, ionotropic, N-methyl D-aspartate 1
|
|
chr19_+_16048306
|
1.892
|
|
TPM4
|
tropomyosin 4
|
|
chr19_+_1892184
|
1.888
|
|
CSNK1G2
|
casein kinase 1, gamma 2
|
|
chr1_+_1104939
|
1.839
|
NM_153254
|
TTLL10
|
tubulin tyrosine ligase-like family, member 10
|
|
chr19_+_1892157
|
1.807
|
NM_001319
|
CSNK1G2
|
casein kinase 1, gamma 2
|
|
chr18_+_22060383
|
1.796
|
NM_005640
|
TAF4B
|
TAF4b RNA polymerase II, TATA box binding protein (TBP)-associated factor, 105kDa
|
|
chr5_-_176760242
|
1.794
|
NM_001029886
|
PFN3
|
profilin 3
|
|
chr11_+_72765044
|
1.763
|
NM_152222
|
RELT
|
RELT tumor necrosis factor receptor
|
|
chr1_+_27591738
|
1.739
|
NM_005281
|
GPR3
|
G protein-coupled receptor 3
|
|
chr8_+_23442307
|
1.737
|
NM_016612
|
SLC25A37
|
solute carrier family 25, member 37
|
|
chr7_+_116447527
|
1.711
|
|
ST7
|
suppression of tumorigenicity 7
|
|
chr19_-_55671273
|
1.702
|
|
FAM71E1
|
family with sequence similarity 71, member E1
|
|
chr1_+_201541269
|
1.695
|
NM_006763
|
BTG2
|
BTG family, member 2
|
|
chr6_+_108593907
|
1.687
|
NM_003269
|
NR2E1
|
nuclear receptor subfamily 2, group E, member 1
|
|
chr1_+_201541241
|
1.682
|
|
BTG2
|
BTG family, member 2
|
|
chr22_+_18499354
|
1.668
|
NM_001185024
NM_013373
|
ZDHHC8
|
zinc finger, DHHC-type containing 8
|
|
chr17_+_24095135
|
1.651
|
NM_004295
|
TRAF4
|
TNF receptor-associated factor 4
|
|
chr10_+_35524835
|
1.642
|
NM_182721
NM_182722
NM_182723
NM_182724
NM_182725
|
CREM
|
cAMP responsive element modulator
|
|
chr11_-_1549719
|
1.637
|
NM_004420
|
DUSP8
|
dual specificity phosphatase 8
|
|
chr7_+_5598964
|
1.620
|
|
FSCN1
|
fascin homolog 1, actin-bundling protein (Strongylocentrotus purpuratus)
|
|
chr11_-_118693021
|
1.620
|
NM_006500
|
MCAM
|
melanoma cell adhesion molecule
|
|
chr9_-_126573349
|
1.615
|
NM_001489
NM_033334
|
NR6A1
|
nuclear receptor subfamily 6, group A, member 1
|
|
chr19_+_54067489
|
1.584
|
|
PPP1R15A
|
protein phosphatase 1, regulatory (inhibitor) subunit 15A
|
|
chr1_+_226394605
|
1.576
|
|
GUK1
|
guanylate kinase 1
|
|
chr19_+_54067450
|
1.575
|
NM_014330
|
PPP1R15A
|
protein phosphatase 1, regulatory (inhibitor) subunit 15A
|
|
chr19_+_18584666
|
1.568
|
NM_012109
|
TMEM59L
|
transmembrane protein 59-like
|
|
chr11_-_69228286
|
1.567
|
NM_005117
|
FGF19
|
fibroblast growth factor 19
|
|
chr16_-_3008162
|
1.566
|
NM_021195
|
CLDN6
|
claudin 6
|
|
chr3_+_188222337
|
1.557
|
NM_003032
|
ST6GAL1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1
|
|
chr7_+_5598979
|
1.550
|
NM_003088
|
FSCN1
|
fascin homolog 1, actin-bundling protein (Strongylocentrotus purpuratus)
|
|
chr6_+_26312758
|
1.539
|
NM_003545
|
HIST1H4E
|
histone cluster 1, H4e
|
|
chr22_+_17539198
|
1.514
|
|
|
|
|
chr1_+_226394589
|
1.510
|
|
GUK1
|
guanylate kinase 1
|
|
chr19_+_1704661
|
1.487
|
NM_001080488
|
ONECUT3
|
one cut homeobox 3
|
|
chr12_-_51734434
|
1.479
|
|
LOC283335
|
hypothetical LOC283335
|
|
chr7_+_30140868
|
1.476
|
NM_152793
|
C7orf41
|
chromosome 7 open reading frame 41
|
|
chr9_-_139473606
|
1.466
|
NM_001130969
NM_001130970
NM_001130971
NM_001178064
NM_015537
|
NELF
|
nasal embryonic LHRH factor
|
|
chr1_+_226394626
|
1.464
|
|
GUK1
|
guanylate kinase 1
|
|
chr19_+_48912053
|
1.460
|
NM_019612
|
IRGC
|
immunity-related GTPase family, cinema
|
|
chr7_-_104696678
|
1.446
|
NM_182691
|
SRPK2
|
SRSF protein kinase 2
|
|
chr5_-_172130766
|
1.441
|
NM_004417
|
DUSP1
|
dual specificity phosphatase 1
|
|
chr19_+_54067491
|
1.441
|
|
PPP1R15A
|
protein phosphatase 1, regulatory (inhibitor) subunit 15A
|
|
chr5_+_174084137
|
1.435
|
NM_002449
|
MSX2
|
msh homeobox 2
|
|
chr19_+_47593137
|
1.434
|
|
|
|
|
chr1_+_226394638
|
1.407
|
|
GUK1
|
guanylate kinase 1
|
|
chr1_+_226394545
|
1.406
|
NM_000858
NM_001159390
|
GUK1
|
guanylate kinase 1
|
|
chr11_-_65094319
|
1.397
|
|
LOC254100
|
hypothetical protein LOC254100
|
|
chr7_+_1239078
|
1.391
|
NM_001080461
|
UNCX
|
UNC homeobox
|
|
chr6_-_15356865
|
1.377
|
|
|
|
|
chr10_-_64246129
|
1.369
|
NM_000399
NM_001136177
NM_001136179
|
EGR2
|
early growth response 2
|
|
chr6_+_15357013
|
1.367
|
|
JARID2
|
jumonji, AT rich interactive domain 2
|
|
chr5_+_118993152
|
1.358
|
NM_001163991
NM_182761
|
FAM170A
|
family with sequence similarity 170, member A
|
|
chr5_-_1935764
|
1.353
|
NM_016358
|
IRX4
|
iroquois homeobox 4
|
|
chr1_-_1274618
|
1.334
|
|
DVL1
|
dishevelled, dsh homolog 1 (Drosophila)
|
|
chr21_-_38792173
|
1.292
|
NM_001136155
NM_182918
|
ERG
|
v-ets erythroblastosis virus E26 oncogene homolog (avian)
|
|
chr9_+_131291144
|
1.273
|
|
|
|
|
chr10_+_35524060
|
1.262
|
NM_182717
NM_182718
NM_182719
NM_182720
|
CREM
|
cAMP responsive element modulator
|
|
chr1_+_110456584
|
1.259
|
NM_203412
|
UBL4B
|
ubiquitin-like 4B
|
|
chr19_+_17047569
|
1.254
|
NM_001130065
NM_004145
|
MYO9B
|
myosin IXB
|
|
chr2_+_232165818
|
1.251
|
NM_152614
|
C2orf57
|
chromosome 2 open reading frame 57
|
|
chr1_-_149955870
|
1.245
|
NM_007185
|
CELF3
|
CUGBP, Elav-like family member 3
|
|
chr11_-_68537243
|
1.230
|
NM_001098515
NM_145015
|
MRGPRF
|
MAS-related GPR, member F
|
|
chr7_-_140270749
|
1.230
|
NM_004333
|
BRAF
|
v-raf murine sarcoma viral oncogene homolog B1
|
|
chr2_+_10969513
|
1.228
|
NM_002236
|
KCNF1
|
potassium voltage-gated channel, subfamily F, member 1
|
|
chr5_-_175897430
|
1.224
|
|
RNF44
|
ring finger protein 44
|
|
chr3_+_42165829
|
1.224
|
|
TRAK1
|
trafficking protein, kinesin binding 1
|
|
chr19_+_55671594
|
1.221
|
|
C19orf63
|
chromosome 19 open reading frame 63
|
|
chr2_+_30307900
|
1.221
|
NM_030915
|
LBH
|
limb bud and heart development homolog (mouse)
|
|
chr3_-_188946168
|
1.219
|
NM_001706
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr19_+_55671468
|
1.207
|
NM_175063
NM_206538
|
C19orf63
|
chromosome 19 open reading frame 63
|
|
chr7_+_128099581
|
1.207
|
NM_001012454
NM_001128926
|
FAM71F2
|
family with sequence similarity 71, member F2
|
|
chr8_-_145983457
|
1.201
|
NM_030580
|
ZNF34
|
zinc finger protein 34
|
|
chrX_-_148857299
|
1.187
|
NM_001013845
|
CXorf40B
|
chromosome X open reading frame 40B
|
|
chr22_+_20326572
|
1.185
|
|
SDF2L1
|
stromal cell-derived factor 2-like 1
|
|
chr19_+_55671600
|
1.184
|
|
C19orf63
|
chromosome 19 open reading frame 63
|
|
chr11_+_76491532
|
1.184
|
NM_006189
|
OMP
|
olfactory marker protein
|
|
chr19_+_13124870
|
1.180
|
|
IER2
|
immediate early response 2
|
|
chr19_-_52308848
|
1.168
|
NM_015168
|
ZC3H4
|
zinc finger CCCH-type containing 4
|
|
chr19_+_55671584
|
1.167
|
|
C19orf63
|
chromosome 19 open reading frame 63
|
|
chr4_-_4300821
|
1.161
|
NM_032927
|
TMEM128
|
transmembrane protein 128
|
|
chr1_+_29086206
|
1.159
|
|
EPB41
|
erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked)
|
|
chr9_-_34387786
|
1.157
|
NM_032596
|
C9orf24
|
chromosome 9 open reading frame 24
|
|
chr8_-_142586487
|
1.156
|
NM_207414
|
FLJ43860
|
FLJ43860 protein
|
|
chr3_-_155524898
|
1.154
|
NM_001114397
NM_020865
|
DHX36
|
DEAH (Asp-Glu-Ala-His) box polypeptide 36
|
|
chr1_+_1207351
|
1.153
|
NM_001130413
NM_002978
|
SCNN1D
|
sodium channel, nonvoltage-gated 1, delta
|
|
chr7_-_158314945
|
1.150
|
|
ESYT2
|
extended synaptotagmin-like protein 2
|
|
chr15_+_29406335
|
1.139
|
NM_015995
|
KLF13
|
Kruppel-like factor 13
|
|
chr19_+_40541327
|
1.132
|
NM_005304
|
FFAR3
|
free fatty acid receptor 3
|
|
chr1_+_19843261
|
1.129
|
NM_005380
|
NBL1
|
neuroblastoma, suppression of tumorigenicity 1
|
|
chr12_+_52634950
|
1.124
|
NM_173860
|
HOXC12
|
homeobox C12
|
|
chr19_-_54231884
|
1.119
|
NM_033377
|
CGB1
CGB2
|
chorionic gonadotropin, beta polypeptide 1
chorionic gonadotropin, beta polypeptide 2
|
|
chr4_-_8481085
|
1.106
|
|
ACOX3
|
acyl-CoA oxidase 3, pristanoyl
|
|
chr19_+_40716153
|
1.105
|
NM_014364
|
GAPDHS
|
glyceraldehyde-3-phosphate dehydrogenase, spermatogenic
|
|
chr11_+_72765318
|
1.086
|
NM_032871
|
RELT
|
RELT tumor necrosis factor receptor
|
|
chr6_+_139497933
|
1.085
|
NM_016217
|
HECA
|
headcase homolog (Drosophila)
|
|
chr9_+_138341752
|
1.080
|
NM_001145638
NM_015597
|
GPSM1
|
G-protein signaling modulator 1
|
|
chr19_+_16048047
|
1.073
|
NM_003290
|
TPM4
|
tropomyosin 4
|
|
chr9_-_139473433
|
1.070
|
|
NELF
|
nasal embryonic LHRH factor
|
|
chr9_+_131291224
|
1.065
|
|
LOC100506190
|
hypothetical LOC100506190
|
|
chr22_+_20326541
|
1.058
|
NM_022044
|
SDF2L1
|
stromal cell-derived factor 2-like 1
|
|
chr11_-_33139449
|
1.047
|
NM_001033505
NM_001033506
NM_001326
|
CSTF3
|
cleavage stimulation factor, 3' pre-RNA, subunit 3, 77kDa
|
|
chr1_+_29086156
|
1.046
|
NM_001166005
NM_001166007
NM_004437
NM_203342
NM_203343
|
EPB41
|
erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked)
|
|
chr1_-_20916789
|
1.044
|
NM_001122819
NM_020816
|
KIF17
|
kinesin family member 17
|
|
chr1_+_36409277
|
1.042
|
|
MAP7D1
|
MAP7 domain containing 1
|
|
chr9_-_139473569
|
1.036
|
|
NELF
|
nasal embryonic LHRH factor
|
|
chr11_+_18676857
|
1.033
|
|
TMEM86A
|
transmembrane protein 86A
|
|
chr3_+_158026769
|
1.028
|
NM_001004316
NM_001193283
|
LEKR1
|
leucine, glutamate and lysine rich 1
|
|
chr22_+_20326591
|
1.014
|
|
SDF2L1
|
stromal cell-derived factor 2-like 1
|
|
chr22_+_20326615
|
1.009
|
|
SDF2L1
|
stromal cell-derived factor 2-like 1
|
|
chr19_+_53801811
|
1.008
|
NM_133498
|
SPACA4
|
sperm acrosome associated 4
|
|
chr20_+_19863716
|
1.008
|
|
RIN2
|
Ras and Rab interactor 2
|
|
chr10_+_99069007
|
0.998
|
NM_005479
|
FRAT1
|
frequently rearranged in advanced T-cell lymphomas
|
|
chr12_+_120635021
|
0.992
|
NM_001080825
|
TMEM120B
|
transmembrane protein 120B
|
|
chr5_+_139907366
|
0.984
|
NM_003732
|
EIF4EBP3
|
eukaryotic translation initiation factor 4E binding protein 3
|
|
chr19_-_50779924
|
0.981
|
NM_001017989
NM_025136
|
OPA3
|
optic atrophy 3 (autosomal recessive, with chorea and spastic paraplegia)
|
|
chr12_-_31368347
|
0.978
|
|
FAM60A
|
family with sequence similarity 60, member A
|
|
chr15_+_74416129
|
0.978
|
|
ISL2
|
ISL LIM homeobox 2
|
|
chr9_+_132990760
|
0.968
|
NM_005085
|
NUP214
|
nucleoporin 214kDa
|
|
chr7_+_100111712
|
0.966
|
|
GNB2
|
guanine nucleotide binding protein (G protein), beta polypeptide 2
|
|
chr16_+_84204424
|
0.965
|
NM_014615
|
KIAA0182
|
KIAA0182
|
|
chr9_-_98421894
|
0.964
|
NM_003671
NM_033331
|
CDC14B
|
CDC14 cell division cycle 14 homolog B (S. cerevisiae)
|
|
chr7_-_5534252
|
0.962
|
|
ACTB
|
actin, beta
|
|
chr2_+_74635354
|
0.961
|
|
DOK1
|
docking protein 1, 62kDa (downstream of tyrosine kinase 1)
|
|
chr4_+_3220564
|
0.955
|
NM_001042690
|
C4orf44
|
chromosome 4 open reading frame 44
|
|
chr3_+_50272648
|
0.942
|
|
|
|
|
chr1_-_226357552
|
0.941
|
NM_024319
|
C1orf35
|
chromosome 1 open reading frame 35
|
|
chr19_-_55671814
|
0.937
|
NM_138411
|
FAM71E1
|
family with sequence similarity 71, member E1
|
|
chr19_-_48791967
|
0.935
|
NM_001007561
|
IRGQ
|
immunity-related GTPase family, Q
|
|
chr19_-_50779914
|
0.926
|
|
OPA3
|
optic atrophy 3 (autosomal recessive, with chorea and spastic paraplegia)
|
|
chr5_+_137829065
|
0.926
|
|
EGR1
|
early growth response 1
|
|
chr16_-_78192111
|
0.916
|
NM_001031804
NM_005360
|
MAF
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog (avian)
|
|
chr21_-_34820916
|
0.911
|
NM_203418
|
RCAN1
|
regulator of calcineurin 1
|
|
chr16_+_4325142
|
0.910
|
|
GLIS2
|
GLIS family zinc finger 2
|
|
chr19_+_53016910
|
0.910
|
NM_000554
|
CRX
|
cone-rod homeobox
|
|
chr5_+_137701122
|
0.906
|
NM_001135647
|
FAM53C
|
family with sequence similarity 53, member C
|
|
chr2_+_96790298
|
0.905
|
NM_020184
|
CNNM4
|
cyclin M4
|
|
chr2_-_74634569
|
0.905
|
NM_032603
|
LOXL3
|
lysyl oxidase-like 3
|
|
chr19_+_13124889
|
0.901
|
|
IER2
|
immediate early response 2
|
|
chr1_-_20685314
|
0.901
|
NM_018584
|
CAMK2N1
|
calcium/calmodulin-dependent protein kinase II inhibitor 1
|
|
chr1_+_242281175
|
0.900
|
NM_205768
|
ZNF238
|
zinc finger protein 238
|
|
chr19_+_50196526
|
0.894
|
NM_006509
|
RELB
|
v-rel reticuloendotheliosis viral oncogene homolog B
|
|
chr13_+_110771015
|
0.887
|
NM_152324
|
C13orf16
|
chromosome 13 open reading frame 16
|
|
chr19_-_53941835
|
0.884
|
NM_182575
|
IZUMO1
|
izumo sperm-egg fusion 1
|
|
chr9_+_101623957
|
0.882
|
NM_006981
NM_173199
|
NR4A3
|
nuclear receptor subfamily 4, group A, member 3
|
|
chr19_+_44591589
|
0.877
|
|
|
|
|
chr22_-_17512097
|
0.876
|
|
DGCR14
|
DiGeorge syndrome critical region gene 14
|
|
chr1_-_10677051
|
0.872
|
|
CASZ1
|
castor zinc finger 1
|
|
chr19_-_50779861
|
0.870
|
|
OPA3
|
optic atrophy 3 (autosomal recessive, with chorea and spastic paraplegia)
|
|
chr19_+_63747678
|
0.868
|
|
TRIM28
|
tripartite motif containing 28
|
|
chr9_-_129517740
|
0.860
|
NM_001002913
|
PTRH1
|
peptidyl-tRNA hydrolase 1 homolog (S. cerevisiae)
|
|
chr11_+_67130898
|
0.859
|
NM_001166102
NM_007103
|
NDUFV1
|
NADH dehydrogenase (ubiquinone) flavoprotein 1, 51kDa
|
|
chr1_+_242281199
|
0.859
|
|
ZNF238
|
zinc finger protein 238
|
|
chr19_-_55706156
|
0.852
|
NM_138334
|
JOSD2
|
Josephin domain containing 2
|
|
chr7_+_116238309
|
0.845
|
|
CAPZA2
|
capping protein (actin filament) muscle Z-line, alpha 2
|
|
chr15_+_66658626
|
0.844
|
NM_006091
|
CORO2B
|
coronin, actin binding protein, 2B
|
|
chr9_+_133154896
|
0.839
|
NM_032728
|
PPAPDC3
|
phosphatidic acid phosphatase type 2 domain containing 3
|
|
chr21_+_46369911
|
0.834
|
|
COL6A2
|
collagen, type VI, alpha 2
|
|
chr11_+_18676913
|
0.832
|
NM_153347
|
TMEM86A
|
transmembrane protein 86A
|
|
chr22_-_17512174
|
0.831
|
NM_022719
|
DGCR14
|
DiGeorge syndrome critical region gene 14
|
|
chr7_-_158314963
|
0.828
|
NM_020728
|
ESYT2
|
extended synaptotagmin-like protein 2
|
|
chr21_+_46369606
|
0.825
|
|
COL6A2
|
collagen, type VI, alpha 2
|
|
chr1_+_207944807
|
0.825
|
NM_005525
|
HSD11B1
|
hydroxysteroid (11-beta) dehydrogenase 1
|
|
chr9_+_129900590
|
0.815
|
|
SLC25A25
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25
|
|
chr15_+_74416156
|
0.813
|
NM_145805
|
ISL2
|
ISL LIM homeobox 2
|
|
chr9_-_129517712
|
0.811
|
|
PTRH1
|
peptidyl-tRNA hydrolase 1 homolog (S. cerevisiae)
|
|
chr17_-_75427644
|
0.791
|
NM_003655
|
CBX4
|
chromobox homolog 4
|
|
chr1_-_243092581
|
0.789
|
|
HNRNPU
|
heterogeneous nuclear ribonucleoprotein U (scaffold attachment factor A)
|
|
chrX_+_153325643
|
0.788
|
NM_004699
|
FAM50A
|
family with sequence similarity 50, member A
|
|
chr12_+_47658502
|
0.785
|
NM_005430
|
WNT1
|
wingless-type MMTV integration site family, member 1
|
|
chr3_-_188945921
|
0.784
|
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr9_-_110658030
|
0.783
|
NM_006686
|
ACTL7B
|
actin-like 7B
|
|
chr10_-_64246080
|
0.781
|
|
EGR2
|
early growth response 2
|
|
chr22_+_18485145
|
0.776
|
|
RANBP1
|
RAN binding protein 1
|
|
chr19_+_60688349
|
0.776
|
NM_020378
|
NAT14
|
N-acetyltransferase 14 (GCN5-related, putative)
|
|
chr3_+_195688974
|
0.774
|
|
FLJ34208
|
hypothetical LOC401106
|
|
chr7_-_5534737
|
0.774
|
|
ACTB
|
actin, beta
|
|
chr5_+_159276317
|
0.771
|
NM_000679
|
ADRA1B
|
adrenergic, alpha-1B-, receptor
|
|
chr7_+_151284396
|
0.771
|
NM_145292
|
GALNTL5
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 5
|
|
chr11_+_116575249
|
0.771
|
NM_001001522
NM_003186
|
TAGLN
|
transgelin
|
|
chr22_+_39404940
|
0.767
|
NM_005297
|
MCHR1
|
melanin-concentrating hormone receptor 1
|