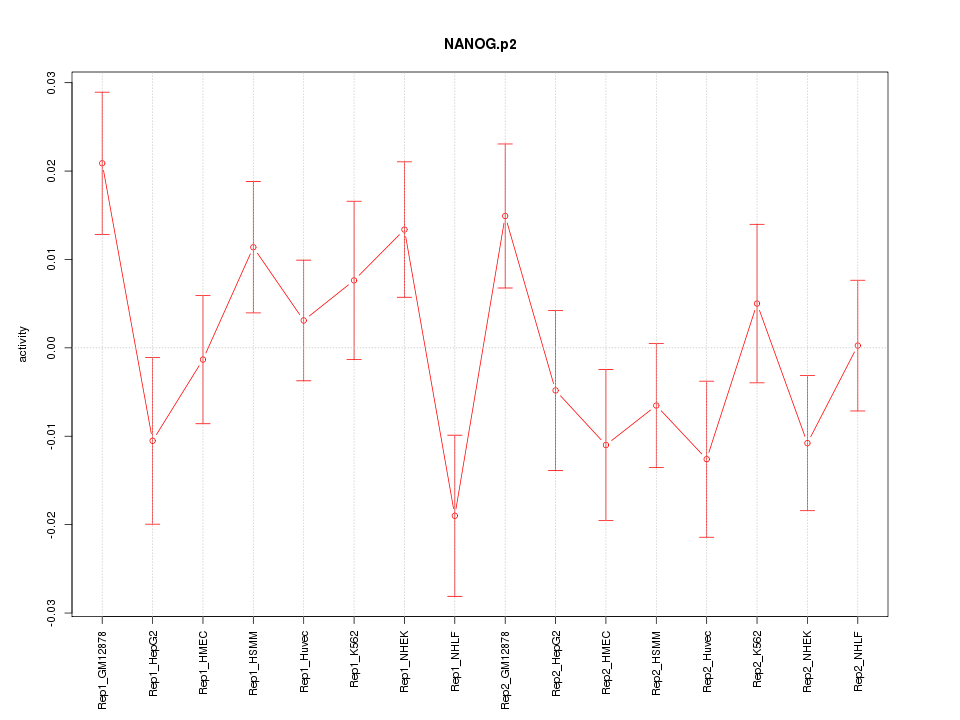
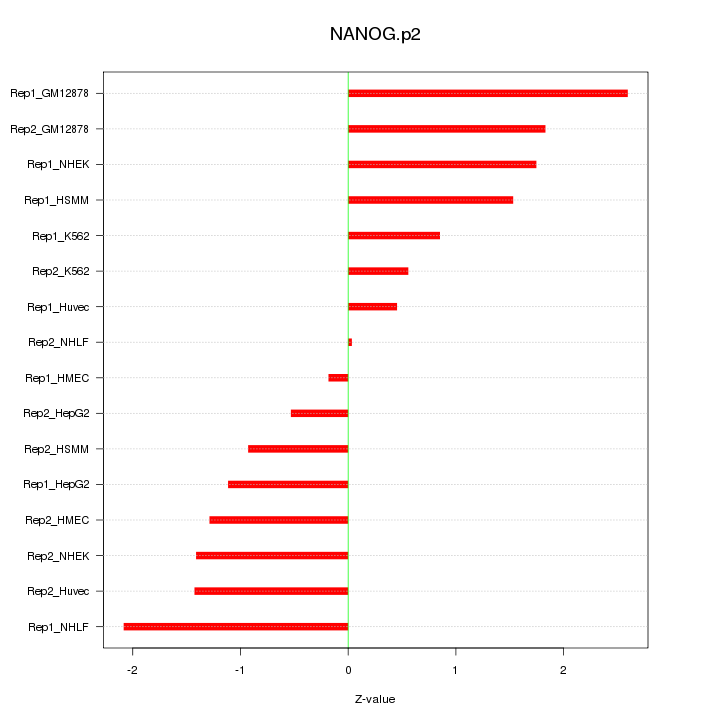
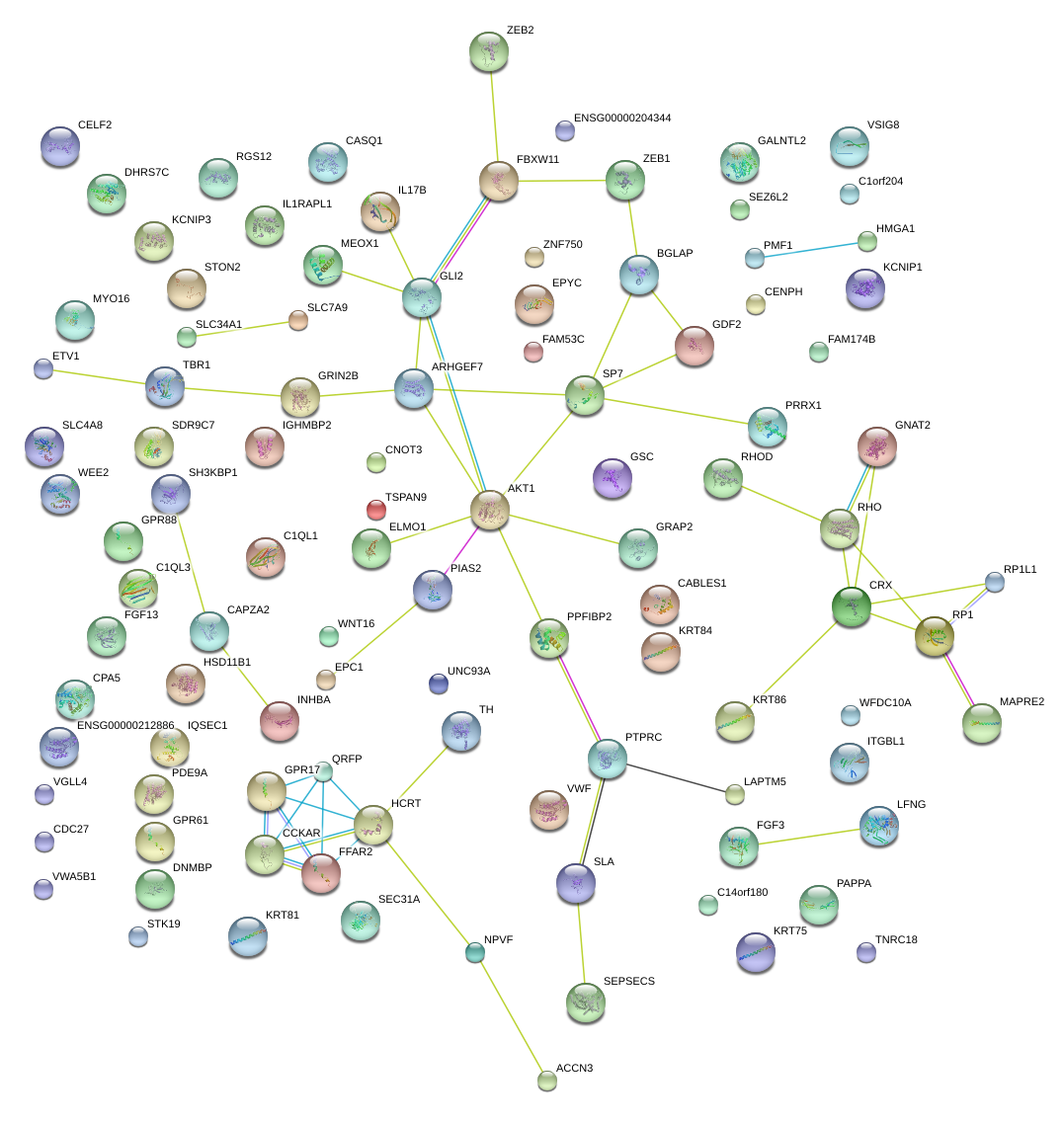
|
chr5_+_169713458
|
2.367
|
NM_001034838
|
KCNIP1
|
Kv channel interacting protein 1
|
|
chr7_-_13992351
|
2.340
|
NM_001163150
NM_001163151
NM_001163152
|
ETV1
|
ets variant 1
|
|
chr6_+_34314545
|
2.238
|
NM_145905
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr1_+_154449389
|
2.068
|
NM_007221
|
PMF1-BGLAP
PMF1
|
PMF1-BGLAP read-through transcript
polyamine-modulated factor 1
|
|
chr3_-_12916547
|
1.989
|
|
IQSEC1
|
IQ motif and Sec7 domain 1
|
|
chr11_-_2149591
|
1.827
|
NM_000360
NM_199292
NM_199293
|
TH
|
tyrosine hydroxylase
|
|
chr1_+_180836050
|
1.612
|
|
|
|
|
chr12_-_51114372
|
1.612
|
NM_004693
|
KRT75
|
keratin 75
|
|
chr19_-_46504831
|
1.511
|
|
|
|
|
chr3_+_130730171
|
1.480
|
NM_000539
|
RHO
|
rhodopsin
|
|
chr7_-_25234582
|
1.470
|
NM_022150
|
NPVF
|
neuropeptide VF precursor
|
|
chr6_+_44076281
|
1.464
|
NM_001171992
NM_153246
|
C6orf223
|
chromosome 6 open reading frame 223
|
|
chr1_+_100776280
|
1.441
|
NM_022049
|
GPR88
|
G protein-coupled receptor 88
|
|
chr12_-_55614311
|
1.384
|
NM_148897
|
SDR9C7
|
short chain dehydrogenase/reductase family 9C, member 7
|
|
chr7_-_96471544
|
1.343
|
|
DLX6-AS1
|
DLX6 antisense RNA 1 (non-protein coding)
|
|
chr9_+_117955890
|
1.337
|
NM_002581
|
PAPPA
|
pregnancy-associated plasma protein A, pappalysin 1
|
|
chr7_+_116238309
|
1.321
|
|
CAPZA2
|
capping protein (actin filament) muscle Z-line, alpha 2
|
|
chr9_-_132759045
|
1.306
|
NM_198180
|
QRFP
|
pyroglutamylated RFamide peptide
|
|
chr12_-_120725210
|
1.293
|
|
LOC338799
|
hypothetical LOC338799
|
|
chr18_+_18989786
|
1.264
|
NM_138375
|
CABLES1
|
Cdk5 and Abl enzyme substrate 1
|
|
chr21_+_43024936
|
1.255
|
|
PDE9A
|
phosphodiesterase 9A
|
|
chr2_+_128119908
|
1.231
|
NM_001161415
NM_001161416
NM_001161417
NM_005291
|
GPR17
|
G protein-coupled receptor 17
|
|
chr7_+_2524022
|
1.217
|
NM_002304
|
LFNG
|
LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase
|
|
chr1_-_30977931
|
1.139
|
|
LAPTM5
|
lysosomal protein transmembrane 5
|
|
chr17_-_9635325
|
1.136
|
NM_001105571
|
DHRS7C
|
dehydrogenase/reductase (SDR family) member 7C
|
|
chr13_+_97856148
|
1.126
|
|
|
|
|
chr13_+_108079570
|
1.103
|
NM_001198950
|
MYO16
|
myosin XVI
|
|
chr3_-_11598829
|
1.100
|
NM_001128220
|
VGLL4
|
vestigial like 4 (Drosophila)
|
|
chr3_+_16191159
|
1.043
|
NM_054110
|
GALNTL2
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 2
|
|
chr11_+_7574992
|
1.034
|
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2)
|
|
chr6_+_167624792
|
1.028
|
NM_001143947
NM_018974
|
UNC93A
|
unc-93 homolog A (C. elegans)
|
|
chr5_-_148738980
|
1.027
|
NM_014443
|
IL17B
|
interleukin 17B
|
|
chr7_+_150376537
|
1.004
|
NM_004769
NM_020321
NM_020322
|
ACCN3
|
amiloride-sensitive cation channel 3
|
|
chr7_+_129771865
|
0.998
|
NM_001127441
NM_001127442
NM_080385
|
CPA5
|
carboxypeptidase A5
|
|
chr14_-_80934523
|
0.994
|
NM_033104
|
STON2
|
stonin 2
|
|
chr1_+_20489998
|
0.984
|
NM_001039500
|
VWA5B1
|
von Willebrand factor A domain containing 5B1
|
|
chr1_+_207944807
|
0.968
|
NM_005525
|
HSD11B1
|
hydroxysteroid (11-beta) dehydrogenase 1
|
|
chr22_+_38652555
|
0.933
|
|
GRAP2
|
GRB2-related adaptor protein 2
|
|
chr4_-_26100986
|
0.903
|
NM_000730
|
CCKAR
|
cholecystokinin A receptor
|
|
chr5_+_176744037
|
0.886
|
NM_001167579
NM_003052
|
SLC34A1
|
solute carrier family 34 (sodium phosphate), member 1
|
|
chr10_-_32707549
|
0.867
|
|
EPC1
|
enhancer of polycomb homolog 1 (Drosophila)
|
|
chr19_-_38052481
|
0.859
|
NM_001126335
NM_014270
|
SLC7A9
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 9
|
|
chrX_-_19727828
|
0.858
|
NM_001024666
|
SH3KBP1
|
SH3-domain kinase binding protein 1
|
|
chr1_-_20682541
|
0.857
|
|
|
|
|
chr19_+_53016910
|
0.853
|
NM_000554
|
CRX
|
cone-rod homeobox
|
|
chr17_-_40401138
|
0.836
|
NM_006688
|
C1QL1
|
complement component 1, q subcomponent-like 1
|
|
chr2_+_161980865
|
0.835
|
NM_006593
|
TBR1
|
T-box, brain, 1
|
|
chr4_+_3341521
|
0.833
|
NM_198227
|
RGS12
|
regulator of G-protein signaling 12
|
|
chr14_-_94306104
|
0.826
|
NM_173849
|
GSC
|
goosecoid homeobox
|
|
chr12_-_14024187
|
0.825
|
NM_000834
|
GRIN2B
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2B
|
|
chr17_-_19290745
|
0.823
|
|
|
|
|
chr12_-_89922888
|
0.815
|
NM_004950
|
EPYC
|
epiphycan
|
|
chr1_+_559618
|
0.808
|
|
|
|
|
chr1_+_196874839
|
0.792
|
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C
|
|
chrX_+_28515479
|
0.791
|
NM_014271
|
IL1RAPL1
|
interleukin 1 receptor accessory protein-like 1
|
|
chr7_-_36992221
|
0.785
|
NM_130442
|
ELMO1
|
engulfment and cell motility 1
|
|
chr7_-_5429702
|
0.772
|
NM_001080495
|
TNRC18
|
trinucleotide repeat containing 18
|
|
chr19_+_59333287
|
0.769
|
|
CNOT3
|
CCR4-NOT transcription complex, subunit 3
|
|
chr15_-_91078307
|
0.768
|
|
FAM174B
|
family with sequence similarity 174, member B
|
|
chr1_+_158426908
|
0.761
|
NM_001231
|
CASQ1
|
calsequestrin 1 (fast-twitch, skeletal muscle)
|
|
chr17_-_37590910
|
0.759
|
NM_001524
|
HCRT
|
hypocretin (orexin) neuropeptide precursor
|
|
chr17_-_78391178
|
0.746
|
NM_024702
|
ZNF750
|
zinc finger protein 750
|
|
chr12_-_52016270
|
0.730
|
NM_001173467
|
SP7
|
Sp7 transcription factor
|
|
chr8_+_55691179
|
0.729
|
NM_006269
|
RP1
|
retinitis pigmentosa 1 (autosomal dominant)
|
|
chrX_-_137649180
|
0.724
|
NM_033642
|
FGF13
|
fibroblast growth factor 13
|
|
chr1_-_158098962
|
0.722
|
NM_001013661
|
VSIG8
|
V-set and immunoglobulin domain containing 8
|
|
chr11_-_69343128
|
0.714
|
NM_005247
|
FGF3
|
fibroblast growth factor 3
|
|
chrX_-_19727775
|
0.702
|
|
SH3KBP1
|
SH3-domain kinase binding protein 1
|
|
chr7_+_120752656
|
0.698
|
NM_016087
|
WNT16
|
wingless-type MMTV integration site family, member 16
|
|
chr14_-_104330982
|
0.685
|
NM_005163
|
AKT1
|
v-akt murine thymoma viral oncogene homolog 1
|
|
chr10_-_48036698
|
0.683
|
NM_016204
|
GDF2
|
growth differentiation factor 2
|
|
chr7_+_141054621
|
0.682
|
NM_001105558
|
WEE2
|
WEE1 homolog 2 (S. pombe)
|
|
chr19_+_40632456
|
0.678
|
NM_005306
|
FFAR2
|
free fatty acid receptor 2
|
|
chr1_-_109957195
|
0.675
|
NM_005272
|
GNAT2
|
guanine nucleotide binding protein (G protein), alpha transducing activity polypeptide 2
|
|
chr10_+_11246934
|
0.673
|
NM_001025076
NM_001083591
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr1_+_109884016
|
0.670
|
NM_031936
|
GPR61
|
G protein-coupled receptor 61
|
|
chr12_-_6103945
|
0.667
|
NM_000552
|
VWF
|
von Willebrand factor
|
|
chr5_+_137701122
|
0.665
|
NM_001135647
|
FAM53C
|
family with sequence similarity 53, member C
|
|
chr2_+_95326753
|
0.652
|
NM_013434
|
KCNIP3
|
Kv channel interacting protein 3, calsenilin
|
|
chr2_-_144994038
|
0.647
|
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr17_-_39094787
|
0.645
|
NM_001040002
|
MEOX1
|
mesenchyme homeobox 1
|
|
chr8_-_10550026
|
0.644
|
NM_178857
|
RP1L1
|
retinitis pigmentosa 1-like 1
|
|
chr12_+_50981915
|
0.641
|
NM_002284
|
KRT86
|
keratin 86
|
|
chr7_-_41709191
|
0.639
|
NM_002192
|
INHBA
|
inhibin, beta A
|
|
chr10_-_101680645
|
0.634
|
|
DNMBP
|
dynamin binding protein
|
|
chr16_-_29817841
|
0.622
|
NM_001114099
NM_001114100
NM_012410
NM_201575
|
SEZ6L2
|
seizure related 6 homolog (mouse)-like 2
|
|
chr13_+_100902942
|
0.619
|
NM_004791
|
ITGBL1
|
integrin, beta-like 1 (with EGF-like repeat domains)
|
|
chr14_+_104117100
|
0.616
|
NM_001008404
|
C14orf180
|
chromosome 14 open reading frame 180
|
|
chr1_+_168898898
|
0.614
|
|
PRRX1
|
paired related homeobox 1
|
|
chr2_+_121271336
|
0.612
|
NM_005270
|
GLI2
|
GLI family zinc finger 2
|
|
chr20_+_43691797
|
0.610
|
NM_080753
|
WFDC10A
|
WAP four-disulfide core domain 10A
|
|
chr12_-_51065629
|
0.603
|
NM_033045
|
KRT84
|
keratin 84
|
|
chr8_-_134156353
|
0.601
|
|
SLA
|
Src-like-adaptor
|
|
chr12_+_25096755
|
0.600
|
|
LRMP
|
lymphoid-restricted membrane protein
|
|
chr1_+_50342168
|
0.599
|
NM_001144776
|
ELAVL4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr12_-_108736983
|
0.599
|
NM_001177431
NM_001177428
NM_001177433
NM_147204
|
TRPV4
|
transient receptor potential cation channel, subfamily V, member 4
|
|
chr3_+_150675117
|
0.599
|
NM_004617
|
TM4SF4
|
transmembrane 4 L six family member 4
|
|
chr17_+_56843893
|
0.598
|
NM_203425
|
C17orf82
|
chromosome 17 open reading frame 82
|
|
chr1_+_196874759
|
0.596
|
NM_002838
NM_080921
NM_080923
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C
|
|
chr4_-_186815116
|
0.593
|
NM_001145674
NM_001145675
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr22_+_38903825
|
0.592
|
NM_001162501
NM_015088
|
TNRC6B
|
trinucleotide repeat containing 6B
|
|
chr8_-_142333339
|
0.590
|
|
SLC45A4
|
solute carrier family 45, member 4
|
|
chr19_-_43926953
|
0.581
|
NM_144691
|
CAPN12
|
calpain 12
|
|
chr9_+_674193
|
0.579
|
|
|
|
|
chr12_-_51233001
|
0.569
|
NM_033448
|
KRT71
|
keratin 71
|
|
chr2_-_40510923
|
0.566
|
NM_001112800
NM_001112801
NM_021097
|
SLC8A1
|
solute carrier family 8 (sodium/calcium exchanger), member 1
|
|
chr4_+_71418886
|
0.564
|
NM_212557
|
AMTN
|
amelotin
|
|
chr6_+_32713160
|
0.558
|
NM_002122
|
HLA-DQA1
|
major histocompatibility complex, class II, DQ alpha 1
|
|
chr12_+_48443232
|
0.556
|
|
TMBIM6
|
transmembrane BAX inhibitor motif containing 6
|
|
chr12_-_89922875
|
0.555
|
|
EPYC
|
epiphycan
|
|
chr7_+_129693938
|
0.555
|
NM_001869
|
CPA2
|
carboxypeptidase A2 (pancreatic)
|
|
chr6_+_34312979
|
0.555
|
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr19_+_7874701
|
0.553
|
|
MAP2K7
|
mitogen-activated protein kinase kinase 7
|
|
chr6_+_34312906
|
0.547
|
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr2_+_157822355
|
0.546
|
NM_014568
|
GALNT5
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 5 (GalNAc-T5)
|
|
chr6_-_154719560
|
0.546
|
NM_001130700
NM_015553
|
IPCEF1
|
interaction protein for cytohesin exchange factors 1
|
|
chr10_+_44726635
|
0.541
|
NM_001123376
|
TMEM72
|
transmembrane protein 72
|
|
chr2_+_161981138
|
0.541
|
|
TBR1
|
T-box, brain, 1
|
|
chr10_-_21475493
|
0.539
|
NM_001010896
NM_001177483
|
C10orf113
|
chromosome 10 open reading frame 113
|
|
chr15_+_56511466
|
0.537
|
NM_000236
|
LIPC
|
lipase, hepatic
|
|
chr5_+_176786279
|
0.533
|
NM_001004105
NM_001004106
NM_002082
|
GRK6
|
G protein-coupled receptor kinase 6
|
|
chr11_+_63775567
|
0.533
|
NM_000932
NM_001184883
|
PLCB3
|
phospholipase C, beta 3 (phosphatidylinositol-specific)
|
|
chr1_-_50661716
|
0.530
|
NM_032110
|
DMRTA2
|
DMRT-like family A2
|
|
chr7_+_18501876
|
0.529
|
NM_014707
NM_178423
|
HDAC9
|
histone deacetylase 9
|
|
chr4_+_71806538
|
0.524
|
NM_001037442
NM_014961
|
RUFY3
|
RUN and FYVE domain containing 3
|
|
chr12_-_51200347
|
0.522
|
NM_000424
|
KRT5
|
keratin 5
|
|
chr3_+_63859114
|
0.520
|
NM_001177387
|
ATXN7
|
ataxin 7
|
|
chr5_+_155686333
|
0.520
|
NM_000337
NM_001128209
NM_172244
|
SGCD
|
sarcoglycan, delta (35kDa dystrophin-associated glycoprotein)
|
|
chr19_+_50663089
|
0.518
|
NM_001114171
NM_006732
|
FOSB
|
FBJ murine osteosarcoma viral oncogene homolog B
|
|
chr6_-_136888771
|
0.512
|
NM_001198608
NM_001198609
NM_001198611
|
MAP7
|
microtubule-associated protein 7
|
|
chr6_+_155579788
|
0.505
|
NM_001010927
|
TIAM2
|
T-cell lymphoma invasion and metastasis 2
|
|
chr5_+_170668892
|
0.499
|
NM_021025
|
TLX3
|
T-cell leukemia homeobox 3
|
|
chr9_+_135491305
|
0.493
|
NM_000787
|
DBH
|
dopamine beta-hydroxylase (dopamine beta-monooxygenase)
|
|
chr12_+_111714096
|
0.492
|
|
RPH3A
|
rabphilin 3A homolog (mouse)
|
|
chrX_-_48713194
|
0.492
|
NM_004979
|
KCND1
|
potassium voltage-gated channel, Shal-related subfamily, member 1
|
|
chr5_+_140703784
|
0.488
|
NM_018916
NM_032011
|
PCDHGA3
|
protocadherin gamma subfamily A, 3
|
|
chr5_+_140509803
|
0.484
|
NM_018939
|
PCDHB6
|
protocadherin beta 6
|
|
chr2_+_161981046
|
0.481
|
|
TBR1
|
T-box, brain, 1
|
|
chr5_+_128468785
|
0.479
|
|
ISOC1
|
isochorismatase domain containing 1
|
|
chr3_-_57209319
|
0.479
|
NM_003865
|
HESX1
|
HESX homeobox 1
|
|
chr17_-_10393664
|
0.476
|
NM_001100112
NM_017534
|
MYH2
|
myosin, heavy chain 2, skeletal muscle, adult
|
|
chr13_-_93929719
|
0.475
|
NM_001129889
NM_001922
|
DCT
|
dopachrome tautomerase (dopachrome delta-isomerase, tyrosine-related protein 2)
|
|
chr7_+_116447527
|
0.472
|
|
ST7
|
suppression of tumorigenicity 7
|
|
chrX_+_21960841
|
0.471
|
NM_000444
|
PHEX
|
phosphate regulating endopeptidase homolog, X-linked
|
|
chr11_+_123986661
|
0.471
|
NM_052959
|
PANX3
|
pannexin 3
|
|
chr11_-_85107569
|
0.463
|
NM_206929
|
SYTL2
|
synaptotagmin-like 2
|
|
chr3_-_55489973
|
0.463
|
|
WNT5A
|
wingless-type MMTV integration site family, member 5A
|
|
chr8_-_134156437
|
0.458
|
|
SLA
|
Src-like-adaptor
|
|
chr2_+_169465995
|
0.454
|
NM_001081686
NM_021176
|
G6PC2
|
glucose-6-phosphatase, catalytic, 2
|
|
chr14_-_56342097
|
0.453
|
NM_172337
|
OTX2
|
orthodenticle homeobox 2
|
|
chr11_-_16380967
|
0.445
|
NM_001145819
NM_017508
|
SOX6
|
SRY (sex determining region Y)-box 6
|
|
chr3_+_69895310
|
0.442
|
NM_001184967
|
MITF
|
microphthalmia-associated transcription factor
|
|
chrX_-_131089552
|
0.440
|
NM_194277
|
FRMD7
|
FERM domain containing 7
|
|
chr7_-_99555399
|
0.439
|
NM_001190415
|
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa
|
|
chr2_-_158439868
|
0.439
|
NM_001105
|
ACVR1
|
activin A receptor, type I
|
|
chr3_+_69895651
|
0.436
|
NM_006722
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr8_+_26569709
|
0.434
|
|
DPYSL2
|
dihydropyrimidinase-like 2
|
|
chr19_+_59333232
|
0.428
|
NM_014516
|
CNOT3
|
CCR4-NOT transcription complex, subunit 3
|
|
chrX_+_70682204
|
0.427
|
|
OGT
|
O-linked N-acetylglucosamine (GlcNAc) transferase (UDP-N-acetylglucosamine:polypeptide-N-acetylglucosaminyl transferase)
|
|
chr9_+_137553104
|
0.427
|
NM_002297
|
LCN1
|
lipocalin 1 (tear prealbumin)
|
|
chrX_+_71280758
|
0.426
|
NM_207422
|
FLJ44635
|
TPT1-like protein
|
|
chr21_+_42583259
|
0.425
|
|
ABCG1
|
ATP-binding cassette, sub-family G (WHITE), member 1
|
|
chr3_-_139315011
|
0.423
|
NM_001170538
|
DZIP1L
|
DAZ interacting protein 1-like
|
|
chr3_-_113842553
|
0.423
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chrX_+_102918409
|
0.422
|
NM_000533
NM_199478
|
PLP1
|
proteolipid protein 1
|
|
chr15_-_53349775
|
0.422
|
NM_183234
NM_004580
|
RAB27A
|
RAB27A, member RAS oncogene family
|
|
chr6_+_50894397
|
0.421
|
NM_003221
|
TFAP2B
|
transcription factor AP-2 beta (activating enhancer binding protein 2 beta)
|
|
chr20_+_36365965
|
0.421
|
NM_001725
|
BPI
|
bactericidal/permeability-increasing protein
|
|
chr15_-_70838703
|
0.416
|
|
|
|
|
chrX_+_102918453
|
0.414
|
|
PLP1
|
proteolipid protein 1
|
|
chr11_-_83071084
|
0.414
|
NM_001142702
|
DLG2
|
discs, large homolog 2 (Drosophila)
|
|
chr10_+_94584449
|
0.413
|
NM_001013848
|
EXOC6
|
exocyst complex component 6
|
|
chr9_+_115303527
|
0.410
|
NM_130795
|
RGS3
|
regulator of G-protein signaling 3
|
|
chr1_+_242283129
|
0.409
|
NM_006352
|
ZNF238
|
zinc finger protein 238
|
|
chr11_-_30558495
|
0.409
|
NM_001584
|
MPPED2
|
metallophosphoesterase domain containing 2
|
|
chr19_+_3713650
|
0.406
|
NM_172251
|
MRPL54
|
mitochondrial ribosomal protein L54
|
|
chr11_+_4427100
|
0.397
|
NM_001005172
|
OR52K2
|
olfactory receptor, family 52, subfamily K, member 2
|
|
chr13_+_26896614
|
0.397
|
NM_002097
|
GTF3A
|
general transcription factor IIIA
|
|
chr1_+_559129
|
0.395
|
|
|
|
|
chr12_-_14740694
|
0.393
|
NM_004963
|
GUCY2C
|
guanylate cyclase 2C (heat stable enterotoxin receptor)
|
|
chr17_-_76720342
|
0.393
|
|
AATK
|
apoptosis-associated tyrosine kinase
|
|
chr1_-_203579932
|
0.390
|
|
KLHDC8A
|
kelch domain containing 8A
|
|
chr12_+_103677388
|
0.389
|
|
CHST11
|
carbohydrate (chondroitin 4) sulfotransferase 11
|
|
chr22_-_16127520
|
0.387
|
|
CECR3
|
cat eye syndrome chromosome region, candidate 3
|
|
chr2_-_180135559
|
0.385
|
NM_001113398
|
ZNF385B
|
zinc finger protein 385B
|
|
chr7_+_150321771
|
0.385
|
NM_001160109
NM_001160110
NM_001160111
|
NOS3
|
nitric oxide synthase 3 (endothelial cell)
|
|
chr8_+_67201684
|
0.385
|
NM_033058
NM_184085
NM_184086
NM_184087
|
TRIM55
|
tripartite motif containing 55
|
|
chrX_+_135397790
|
0.383
|
NM_001727
|
BRS3
|
bombesin-like receptor 3
|
|
chr3_-_11585264
|
0.382
|
NM_001128221
|
VGLL4
|
vestigial like 4 (Drosophila)
|
|
chr11_+_130745580
|
0.382
|
NM_001048209
|
NTM
|
neurotrimin
|
|
chr12_-_51457371
|
0.381
|
NM_015848
|
KRT76
|
keratin 76
|
|
chr6_-_116973465
|
0.381
|
NM_001139444
|
BET3L
|
BET3 like (S. cerevisiae)
|
|
chr7_+_113842284
|
0.377
|
NM_001172766
NM_014491
NM_148898
NM_148900
|
FOXP2
|
forkhead box P2
|
|
chr5_+_60969392
|
0.377
|
NM_173667
|
FLJ37543
|
hypothetical protein FLJ37543
|
|
chr8_-_42353719
|
0.376
|
NM_014420
|
DKK4
|
dickkopf homolog 4 (Xenopus laevis)
|
|
chr22_-_37211467
|
0.376
|
|
DDX17
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 17
|
|
chr19_+_59973074
|
0.375
|
NM_014218
|
KIR2DS2
KIR2DL1
|
killer cell immunoglobulin-like receptor, two domains, short cytoplasmic tail, 2
killer cell immunoglobulin-like receptor, two domains, long cytoplasmic tail, 1
|
|
chr14_+_77939845
|
0.374
|
NM_004796
|
NRXN3
|
neurexin 3
|
|
chr3_+_157877146
|
0.374
|
NM_001184718
|
TIPARP
|
TCDD-inducible poly(ADP-ribose) polymerase
|
|
chr1_+_151456683
|
0.374
|
NM_001195571
|
PRR9
|
proline rich 9
|