|
chr22_+_21484240
|
8.890
|
|
|
|
|
chr22_+_21340757
|
7.620
|
|
|
|
|
chr22_+_21359175
|
4.638
|
|
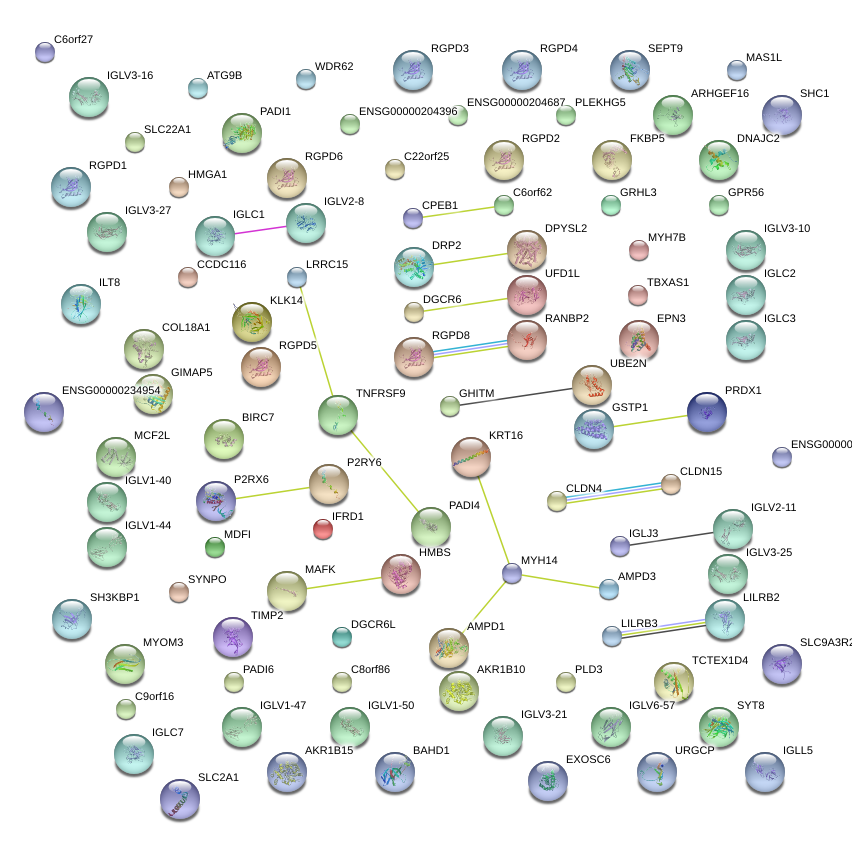
IGLV3-25
|
immunoglobulin lambda variable 3-25
|
|
chr22_+_21370394
|
4.509
|
|
IGL@
|
immunoglobulin lambda locus
|
|
chr22_+_21495273
|
3.803
|
|
|
|
|
chr22_+_21079355
|
3.350
|
|
|
|
|
chr22_+_21053972
|
3.345
|
|
|
|
|
chr22_+_21407066
|
3.246
|
|
|
|
|
chr22_+_21393107
|
3.134
|
|
IGLJ3
|
immunoglobulin lambda joining 3
|
|
chr22_+_20899155
|
3.037
|
|
|
|
|
chr1_-_24307514
|
2.755
|
|
MYOM3
|
myomesin family, member 3
|
|
chr9_+_129962292
|
2.568
|
NM_024112
|
C9orf16
|
chromosome 9 open reading frame 16
|
|
chr22_+_21094094
|
2.392
|
|
|
|
|
chr22_+_21116283
|
2.260
|
|
|
|
|
chr22_+_18388656
|
2.253
|
|
C22orf25
|
chromosome 22 open reading frame 25
|
|
chr22_+_21042091
|
2.252
|
|
IGLV1-44
IGL@
|
immunoglobulin lambda variable 1-44
immunoglobulin lambda locus
|
|
chr22_+_21559959
|
2.047
|
NM_001178126
|
IGLL5
IGLC1
|
immunoglobulin lambda-like polypeptide 5
immunoglobulin lambda constant 1 (Mcg marker)
|
|
chr22_+_20316968
|
2.042
|
NM_152612
|
CCDC116
|
coiled-coil domain containing 116
|
|
chr1_+_24522116
|
1.880
|
NM_021180
|
GRHL3
|
grainyhead-like 3 (Drosophila)
|
|
chr22_+_21065134
|
1.823
|
|
LOC100290481
|
immunoglobulin lambda light chain-like
|
|
chr22_+_21006814
|
1.774
|
|
|
|
|
chr1_-_6474346
|
1.743
|
NM_020631
|
PLEKHG5
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5
|
|
chr22_+_21552897
|
1.707
|
|
LOC100290481
|
immunoglobulin lambda light chain-like
|
|
chr22_+_21065207
|
1.703
|
|
IGLJ3
|
immunoglobulin lambda joining 3
|
|
chr22_-_17846711
|
1.659
|
|
UFD1L
|
ubiquitin fusion degradation 1 like (yeast)
|
|
chr22_-_17846621
|
1.654
|
|
UFD1L
|
ubiquitin fusion degradation 1 like (yeast)
|
|
chr22_+_21065202
|
1.651
|
|
|
|
|
chr22_+_19699441
|
1.589
|
NM_001159554
NM_005446
|
P2RX6
|
purinergic receptor P2X, ligand-gated ion channel, 6
|
|
chr6_+_34314545
|
1.582
|
NM_145905
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr1_+_17571277
|
1.541
|
NM_207421
|
PADI6
|
peptidyl arginine deiminase, type VI
|
|
chr1_+_3378030
|
1.533
|
|
ARHGEF16
|
Rho guanine nucleotide exchange factor (GEF) 16
|
|
chr22_+_21011653
|
1.499
|
|
|
|
|
chr22_-_17846725
|
1.478
|
NM_001035247
NM_005659
|
UFD1L
|
ubiquitin fusion degradation 1 like (yeast)
|
|
chr11_+_118463906
|
1.462
|
NM_001024382
|
HMBS
|
hydroxymethylbilane synthase
|
|
chr22_+_20880159
|
1.454
|
|
IGLV6-57
|
immunoglobulin lambda variable 6-57
|
|
chr7_-_150561233
|
1.390
|
|
|
|
|
chr1_-_45760115
|
1.384
|
NM_002574
NM_181696
NM_181697
|
PRDX1
|
peroxiredoxin 1
|
|
chr21_+_45699830
|
1.374
|
NM_030582
NM_130444
|
COL18A1
|
collagen, type XVIII, alpha 1
|
|
chr7_+_1536847
|
1.369
|
NM_002360
|
MAFK
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog K (avian)
|
|
chr16_+_2023279
|
1.363
|
|
SLC9A3R2
|
solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 2
|
|
chr17_-_37022465
|
1.236
|
NM_005557
|
KRT16
|
keratin 16
|
|
chr2_+_87536174
|
1.235
|
|
NCRNA00152
|
non-protein coding RNA 152
|
|
chr6_-_31853027
|
1.224
|
NM_025258
|
C6orf27
|
chromosome 6 open reading frame 27
|
|
chr2_+_87536106
|
1.208
|
|
NCRNA00152
|
non-protein coding RNA 152
|
|
chr11_+_1812115
|
1.207
|
NM_138567
|
SYT8
|
synaptotagmin VIII
|
|
chr1_+_17404206
|
1.201
|
NM_013358
|
PADI1
|
peptidyl arginine deiminase, type I
|
|
chr7_+_150065594
|
1.184
|
|
GIMAP5
|
GTPase, IMAP family member 5
|
|
chr7_+_1543644
|
1.172
|
|
MAFK
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog K (avian)
|
|
chr10_+_85889164
|
1.091
|
NM_014394
|
GHITM
|
growth hormone inducible transmembrane protein
|
|
chr3_-_195571748
|
1.075
|
NM_001135057
NM_130830
|
LRRC15
|
leucine rich repeat containing 15
|
|
chr7_-_100667750
|
1.066
|
NM_014343
|
CLDN15
|
claudin 15
|
|
chr15_-_81037554
|
1.039
|
NM_001079533
NM_001079534
NM_001079535
|
CPEB1
|
cytoplasmic polyadenylation element binding protein 1
|
|
chr7_+_72883128
|
1.020
|
NM_001305
|
CLDN4
|
claudin 4
|
|
chr5_+_150000394
|
1.004
|
NM_001109974
NM_007286
|
SYNPO
|
synaptopodin
|
|
chr7_+_139175420
|
0.974
|
NM_001061
NM_001166253
NM_030984
|
TBXAS1
|
thromboxane A synthase 1 (platelet)
|
|
chr1_-_153209625
|
0.945
|
NM_001130040
NM_183001
|
SHC1
|
SHC (Src homology 2 domain containing) transforming protein 1
|
|
chr7_+_150065376
|
0.929
|
NM_018384
|
GIMAP5
|
GTPase, IMAP family member 5
|
|
chr16_+_56219802
|
0.914
|
NM_001145771
NM_001145772
NM_001145773
NM_201524
NM_201525
|
GPR56
|
G protein-coupled receptor 56
|
|
chr16_-_68843295
|
0.913
|
NM_058219
|
EXOSC6
|
exosome component 6
|
|
chr19_-_56279278
|
0.910
|
NM_022046
|
KLK14
|
kallikrein-related peptidase 14
|
|
chr6_-_29563657
|
0.909
|
NM_052967
|
MAS1L
|
MAS1 oncogene-like
|
|
chr1_-_45045543
|
0.902
|
NM_001013632
|
TCTEX1D4
|
Tctex1 domain containing 4
|
|
chr22_+_21027538
|
0.886
|
|
|
|
|
chr22_+_17273735
|
0.885
|
NM_005675
|
DGCR6
|
DiGeorge syndrome critical region gene 6
|
|
chr6_+_160462852
|
0.874
|
NM_003057
NM_153187
|
SLC22A1
|
solute carrier family 22 (organic cation transporter), member 1
|
|
chr15_+_38520594
|
0.853
|
NM_014952
|
BAHD1
|
bromo adjacent homology domain containing 1
|
|
chr1_-_115039614
|
0.852
|
NM_000036
NM_001172626
|
AMPD1
|
adenosine monophosphate deaminase 1
|
|
chr7_-_150352484
|
0.848
|
NM_173681
|
ATG9B
|
ATG9 autophagy related 9 homolog B (S. cerevisiae)
|
|
chr7_+_111850367
|
0.847
|
NM_001007245
NM_001197080
|
IFRD1
|
interferon-related developmental regulator 1
|
|
chr1_+_17507276
|
0.846
|
NM_012387
|
PADI4
|
peptidyl arginine deiminase, type IV
|
|
chrX_+_100361588
|
0.842
|
NM_001171184
NM_001939
|
DRP2
|
dystrophin related protein 2
|
|
chr6_-_35804337
|
0.830
|
NM_001145775
|
FKBP5
|
FK506 binding protein 5
|
|
chr22_+_20880112
|
0.828
|
|
|
|
|
chr6_+_41712907
|
0.819
|
|
MDFI
|
MyoD family inhibitor
|
|
chr17_+_45965076
|
0.781
|
|
EPN3
|
epsin 3
|
|
chr11_+_72660894
|
0.778
|
NM_004154
|
P2RY6
|
pyrimidinergic receptor P2Y, G-protein coupled, 6
|
|
chr7_+_133862883
|
0.774
|
NM_020299
|
AKR1B10
|
aldo-keto reductase family 1, member B10 (aldose reductase)
|
|
chr17_-_74433048
|
0.771
|
NM_003255
|
TIMP2
|
TIMP metallopeptidase inhibitor 2
|
|
chr13_+_112746908
|
0.769
|
|
MCF2L
|
MCF.2 cell line derived transforming sequence-like
|
|
chr1_-_7923464
|
0.768
|
NM_001561
|
TNFRSF9
|
tumor necrosis factor receptor superfamily, member 9
|
|
chr19_+_45564549
|
0.765
|
|
PLD3
|
phospholipase D family, member 3
|
|
chr8_-_38505331
|
0.756
|
NM_207412
|
C8orf86
|
chromosome 8 open reading frame 86
|
|
chr17_-_74432800
|
0.755
|
|
TIMP2
|
TIMP metallopeptidase inhibitor 2
|
|
chr19_-_59438411
|
0.750
|
NM_024318
|
LILRB3
LILRA6
|
leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 3
leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 6
|
|
chr19_+_55398680
|
0.741
|
NM_001077186
NM_001145809
NM_024729
|
MYH14
|
myosin, heavy chain 14, non-muscle
|
|
chrX_-_19598912
|
0.740
|
NM_001184960
|
SH3KBP1
|
SH3-domain kinase binding protein 1
|
|
chr20_+_61337679
|
0.738
|
NM_022161
NM_139317
|
BIRC7
|
baculoviral IAP repeat containing 7
|
|
chr11_+_10433241
|
0.726
|
NM_001025389
|
AMPD3
|
adenosine monophosphate deaminase 3
|
|
chr7_-_102772304
|
0.718
|
NM_001129887
NM_014377
|
DNAJC2
|
DnaJ (Hsp40) homolog, subfamily C, member 2
|
|
chr11_+_67107852
|
0.707
|
|
GSTP1
|
glutathione S-transferase pi 1
|
|
chr1_-_43168247
|
0.707
|
|
SLC2A1
|
solute carrier family 2 (facilitated glucose transporter), member 1
|
|
chr2_-_112907577
|
0.706
|
NM_001164463
|
RGPD8
RGPD5
|
RANBP2-like and GRIP domain containing 8
RANBP2-like and GRIP domain containing 5
|
|
chr6_-_24827265
|
0.700
|
NM_030939
|
C6orf62
|
chromosome 6 open reading frame 62
|
|
chr19_-_54212158
|
0.694
|
NM_000894
|
LHB
|
luteinizing hormone beta polypeptide
|
|
chr7_+_130445394
|
0.683
|
NM_001145354
|
MKLN1
|
muskelin 1, intracellular mediator containing kelch motifs
|
|
chr16_-_66075216
|
0.681
|
NM_001138
|
AGRP
|
agouti related protein homolog (mouse)
|
|
chr16_+_1230678
|
0.678
|
NM_003294
|
TPSB2
TPSAB1
|
tryptase beta 2 (gene/pseudogene)
tryptase alpha/beta 1
|
|
chr21_+_38566298
|
0.668
|
NM_170737
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15
|
|
chr6_-_28429950
|
0.663
|
NM_145909
|
ZNF323
|
zinc finger protein 323
|
|
chr17_+_71109734
|
0.663
|
|
MYO15B
|
myosin XVB pseudogene
|
|
chr1_+_27591738
|
0.661
|
NM_005281
|
GPR3
|
G protein-coupled receptor 3
|
|
chr3_+_46894239
|
0.649
|
NM_000316
|
PTH1R
|
parathyroid hormone 1 receptor
|
|
chr19_+_3317589
|
0.646
|
|
NFIC
|
nuclear factor I/C (CCAAT-binding transcription factor)
|
|
chr7_+_150319076
|
0.643
|
NM_000603
|
NOS3
|
nitric oxide synthase 3 (endothelial cell)
|
|
chr12_-_46405477
|
0.635
|
NM_001172439
NM_001172440
NM_006025
|
ENDOU
|
endonuclease, polyU-specific
|
|
chr12_-_49897743
|
0.623
|
|
POU6F1
|
POU class 6 homeobox 1
|
|
chr11_+_67107641
|
0.620
|
NM_000852
|
GSTP1
|
glutathione S-transferase pi 1
|
|
chr6_+_43373986
|
0.618
|
|
SLC22A7
|
solute carrier family 22 (organic anion transporter), member 7
|
|
chrX_+_12903145
|
0.613
|
NM_021109
NM_183049
|
TMSB4X
TMSL3
|
thymosin beta 4, X-linked
thymosin-like 3
|
|
chr1_+_158426908
|
0.611
|
NM_001231
|
CASQ1
|
calsequestrin 1 (fast-twitch, skeletal muscle)
|
|
chrX_-_109570113
|
0.609
|
NM_001171689
|
AMMECR1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1
|
|
chr10_-_76538859
|
0.598
|
NM_001007271
NM_001007272
NM_001007273
|
DUSP13
|
dual specificity phosphatase 13
|
|
chr7_+_129720209
|
0.597
|
NM_001163446
NM_016352
|
CPA4
|
carboxypeptidase A4
|
|
chr1_-_115682374
|
0.595
|
NM_002506
|
NGF
|
nerve growth factor (beta polypeptide)
|
|
chr11_+_62136788
|
0.592
|
NM_000327
|
ROM1
|
retinal outer segment membrane protein 1
|
|
chrX_-_48713194
|
0.583
|
NM_004979
|
KCND1
|
potassium voltage-gated channel, Shal-related subfamily, member 1
|
|
chr19_+_3317575
|
0.577
|
|
NFIC
|
nuclear factor I/C (CCAAT-binding transcription factor)
|
|
chr8_-_143820825
|
0.571
|
NM_020427
|
SLURP1
|
secreted LY6/PLAUR domain containing 1
|
|
chr16_-_27806978
|
0.570
|
NM_144675
|
GSG1L
|
GSG1-like
|
|
chr11_-_62136778
|
0.568
|
NM_153265
|
EML3
|
echinoderm microtubule associated protein like 3
|
|
chr6_+_31190505
|
0.558
|
NM_014068
|
PSORS1C1
|
psoriasis susceptibility 1 candidate 1
|
|
chr19_+_56319948
|
0.556
|
NM_001198558
NM_014441
|
SIGLEC9
|
sialic acid binding Ig-like lectin 9
|
|
chr11_-_6298284
|
0.554
|
NM_145040
|
PRKCDBP
|
protein kinase C, delta binding protein
|
|
chr1_+_36327039
|
0.550
|
NM_017825
|
ADPRHL2
|
ADP-ribosylhydrolase like 2
|
|
chr3_-_152403636
|
0.548
|
NM_013308
|
GPR171
|
G protein-coupled receptor 171
|
|
chr8_-_42742729
|
0.547
|
NM_004198
|
CHRNA6
|
cholinergic receptor, nicotinic, alpha 6
|
|
chr7_+_1050704
|
0.540
|
|
|
|
|
chr16_+_82544327
|
0.540
|
NM_182981
|
OSGIN1
|
oxidative stress induced growth inhibitor 1
|
|
chr1_+_19511326
|
0.534
|
NM_001040125
NM_001040126
NM_017765
|
PQLC2
|
PQ loop repeat containing 2
|
|
chr22_-_18687511
|
0.529
|
NM_033257
|
DGCR6L
|
DiGeorge syndrome critical region gene 6-like
|
|
chr7_+_106991638
|
0.526
|
NM_181581
|
DUS4L
|
dihydrouridine synthase 4-like (S. cerevisiae)
|
|
chr19_-_43955984
|
0.522
|
NM_002307
|
LGALS7
LGALS7B
|
lectin, galactoside-binding, soluble, 7
lectin, galactoside-binding, soluble, 7B
|
|
chr7_-_75280962
|
0.522
|
NM_002991
|
CCL24
|
chemokine (C-C motif) ligand 24
|
|
chr6_-_154719560
|
0.510
|
NM_001130700
NM_015553
|
IPCEF1
|
interaction protein for cytohesin exchange factors 1
|
|
chr11_-_116605761
|
0.507
|
|
PCSK7
|
proprotein convertase subtilisin/kexin type 7
|
|
chr19_+_56337368
|
0.505
|
NM_014385
NM_016543
|
SIGLEC7
|
sialic acid binding Ig-like lectin 7
|
|
chrX_+_153423652
|
0.505
|
NM_001099856
|
IKBKG
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma
|
|
chr11_+_64079665
|
0.501
|
NM_018484
|
SLC22A11
|
solute carrier family 22 (organic anion/urate transporter), member 11
|
|
chr19_-_8118367
|
0.493
|
NM_032447
|
FBN3
|
fibrillin 3
|
|
chr3_+_130730171
|
0.491
|
NM_000539
|
RHO
|
rhodopsin
|
|
chr22_+_17498320
|
0.485
|
NM_053006
|
TSSK2
|
testis-specific serine kinase 2
|
|
chr4_-_38710376
|
0.484
|
NM_024943
|
TMEM156
|
transmembrane protein 156
|
|
chr6_-_170704251
|
0.482
|
|
PSMB1
|
proteasome (prosome, macropain) subunit, beta type, 1
|
|
chr7_+_157339985
|
0.482
|
|
LOC100506585
|
hypothetical LOC100506585
|
|
chr11_+_74548472
|
0.481
|
NM_001145211
|
SLCO2B1
|
solute carrier organic anion transporter family, member 2B1
|
|
chr7_+_142768916
|
0.478
|
|
ZYX
|
zyxin
|
|
chr15_+_32304477
|
0.469
|
NM_016454
|
TMEM85
|
transmembrane protein 85
|
|
chr21_+_31953803
|
0.468
|
NM_000454
|
SOD1
|
superoxide dismutase 1, soluble
|
|
chr22_+_18090118
|
0.463
|
|
SEPT5-GP1BB
|
SEPT5-GP1BB read-through transcript
|
|
chr1_+_153232685
|
0.461
|
NM_018655
|
LENEP
|
lens epithelial protein
|
|
chr5_-_75954807
|
0.460
|
NM_004101
|
F2RL2
|
coagulation factor II (thrombin) receptor-like 2
|
|
chr9_-_139042526
|
0.456
|
NM_001606
|
ABCA2
|
ATP-binding cassette, sub-family A (ABC1), member 2
|
|
chr1_-_45249603
|
0.456
|
NM_024602
|
HECTD3
|
HECT domain containing 3
|
|
chr10_-_73167586
|
0.452
|
NM_001168390
|
C10orf105
|
chromosome 10 open reading frame 105
|
|
chr12_+_63084317
|
0.448
|
NM_007235
|
XPOT
|
exportin, tRNA (nuclear export receptor for tRNAs)
|
|
chr15_-_71830868
|
0.448
|
NM_001039614
|
C15orf59
|
chromosome 15 open reading frame 59
|
|
chr19_-_40711034
|
0.448
|
NM_001166034
NM_001166035
NM_198538
|
SBSN
|
suprabasin
|
|
chr15_+_39638445
|
0.445
|
NM_006293
|
TYRO3
|
TYRO3 protein tyrosine kinase
|
|
chrX_+_133198742
|
0.435
|
NM_001101357
|
CCDC160
|
coiled-coil domain containing 160
|
|
chr1_-_33139425
|
0.433
|
NM_033504
|
TMEM54
|
transmembrane protein 54
|
|
chr19_+_3317536
|
0.432
|
NM_005597
|
NFIC
|
nuclear factor I/C (CCAAT-binding transcription factor)
|
|
chr9_-_139042416
|
0.432
|
|
ABCA2
|
ATP-binding cassette, sub-family A (ABC1), member 2
|
|
chr21_+_46367216
|
0.422
|
|
COL6A2
|
collagen, type VI, alpha 2
|
|
chr8_+_22266256
|
0.421
|
|
PIWIL2
|
piwi-like 2 (Drosophila)
|
|
chr9_+_139255559
|
0.420
|
|
TUBB2C
|
tubulin, beta 2C
|
|
chrX_-_100527799
|
0.417
|
NM_000061
|
BTK
|
Bruton agammaglobulinemia tyrosine kinase
|
|
chr2_+_173794692
|
0.417
|
|
ZAK
|
sterile alpha motif and leucine zipper containing kinase AZK
|
|
chr11_-_62539857
|
0.415
|
NM_001184732
NM_001184733
NM_001184736
NM_004254
|
SLC22A8
|
solute carrier family 22 (organic anion transporter), member 8
|
|
chr22_-_43040063
|
0.413
|
NM_001099294
|
KIAA1644
|
KIAA1644
|
|
chr1_+_46578976
|
0.413
|
NM_199044
|
NSUN4
|
NOP2/Sun domain family, member 4
|
|
chr19_-_40696393
|
0.406
|
NM_001126056
NM_001126057
NM_001126058
NM_001190347
NM_001190348
NM_001190349
NM_033317
|
DMKN
|
dermokine
|
|
chr6_-_3102690
|
0.403
|
NM_001069
|
TUBB2A
|
tubulin, beta 2A
|
|
chr19_+_991079
|
0.403
|
NM_019112
|
ABCA7
|
ATP-binding cassette, sub-family A (ABC1), member 7
|
|
chr2_+_113591940
|
0.403
|
NM_000577
NM_173841
NM_173843
|
IL1RN
|
interleukin 1 receptor antagonist
|
|
chr10_+_75202258
|
0.401
|
|
FUT11
|
fucosyltransferase 11 (alpha (1,3) fucosyltransferase)
|
|
chr1_-_84237316
|
0.398
|
NM_024686
|
TTLL7
|
tubulin tyrosine ligase-like family, member 7
|
|
chr6_-_30793436
|
0.397
|
NM_014641
|
MDC1
|
mediator of DNA-damage checkpoint 1
|
|
chr9_+_139255519
|
0.395
|
NM_006088
|
TUBB2C
|
tubulin, beta 2C
|
|
chr11_-_64906683
|
0.393
|
NM_001077241
NM_182556
|
SLC25A45
|
solute carrier family 25, member 45
|
|
chr1_-_153195219
|
0.388
|
|
PBXIP1
|
pre-B-cell leukemia homeobox interacting protein 1
|
|
chr10_+_75202071
|
0.388
|
|
FUT11
|
fucosyltransferase 11 (alpha (1,3) fucosyltransferase)
|
|
chr7_-_115458047
|
0.387
|
|
TFEC
|
transcription factor EC
|
|
chr7_-_33047042
|
0.386
|
NM_001166118
NM_016489
|
NT5C3
|
5'-nucleotidase, cytosolic III
|
|
chr6_-_2580296
|
0.383
|
NM_152554
|
C6orf195
|
chromosome 6 open reading frame 195
|
|
chr1_-_45249554
|
0.382
|
|
HECTD3
|
HECT domain containing 3
|
|
chr1_-_111908085
|
0.380
|
NM_001081976
|
ADORA3
|
adenosine A3 receptor
|
|
chr6_+_31782618
|
0.380
|
NM_001003693
|
LY6G6F
|
lymphocyte antigen 6 complex, locus G6F
|
|
chr9_+_94776538
|
0.377
|
NM_033086
|
FGD3
|
FYVE, RhoGEF and PH domain containing 3
|
|
chr9_+_131291224
|
0.377
|
|
LOC100506190
|
hypothetical LOC100506190
|
|
chr6_-_32929721
|
0.377
|
NM_000593
|
TAP1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP)
|
|
chr16_+_28413421
|
0.377
|
NM_018690
|
APOBR
|
apolipoprotein B receptor
|
|
chr9_+_131291144
|
0.374
|
|
|
|
|
chr1_-_43523825
|
0.373
|
NM_182517
NM_001164829
|
C1orf210
|
chromosome 1 open reading frame 210
|
|
chr9_-_99974994
|
0.373
|
NM_003389
|
CORO2A
|
coronin, actin binding protein, 2A
|
|
chr16_+_87765661
|
0.372
|
NM_004933
|
CDH15
|
cadherin 15, type 1, M-cadherin (myotubule)
|
|
chr6_-_33655995
|
0.362
|
NM_001188
|
BAK1
|
BCL2-antagonist/killer 1
|
|
chr3_+_5204430
|
0.361
|
|
EDEM1
|
ER degradation enhancer, mannosidase alpha-like 1
|
|
chr7_-_76667045
|
0.361
|
NM_006682
|
FGL2
|
fibrinogen-like 2
|
|
chr2_+_74635019
|
0.358
|
NM_001381
|
DOK1
|
docking protein 1, 62kDa (downstream of tyrosine kinase 1)
|
|
chr1_-_240228987
|
0.358
|
NM_001004343
|
MAP1LC3C
|
microtubule-associated protein 1 light chain 3 gamma
|